bioinformatics @Twist
formerly: bioinformatics lead at plasmidsaurus, PhD at University of Texas at Austin
bioinformatics, bacteriology, phages, engineered plasmids, synthetic biology, insects
I wrote a short piece reflecting on the discovery and significance of this metric (and really enjoyed digging into the context and story behind it!) #microsky 🧬
I wrote a short piece reflecting on the discovery and significance of this metric (and really enjoyed digging into the context and story behind it!) #microsky 🧬

Ie: it's not us (Samtools team)! Be warned
Ie: it's not us (Samtools team)! Be warned
- Rich CIGAR and base visualization
- Allele frequency visualization
- VCF and BED file support
- Mouse dragging and hovering
- Filter alignment
Now 90% of what I need from IGV can be done in the terminal.
Some interesting behind-the-scenes:
- Rich CIGAR and base visualization
- Allele frequency visualization
- VCF and BED file support
- Mouse dragging and hovering
- Filter alignment
Now 90% of what I need from IGV can be done in the terminal.
Some interesting behind-the-scenes:
@yanwong.bsky.social and I have been developing a method to programmatically drawing these graphs.
🔗 arxiv.org/abs/2508.03958
1/6

@yanwong.bsky.social and I have been developing a method to programmatically drawing these graphs.
🔗 arxiv.org/abs/2508.03958
1/6
genomebiology.biomedcentral.com/articles/10....

genomebiology.biomedcentral.com/articles/10....
Hidden microbial world in trees🌳
Living wood hosts trillions of bacteria making trees a complex ecosystems with major roles in forest health and function.
#PlantMicro #MicroSky
www.nature.com/articles/s41...
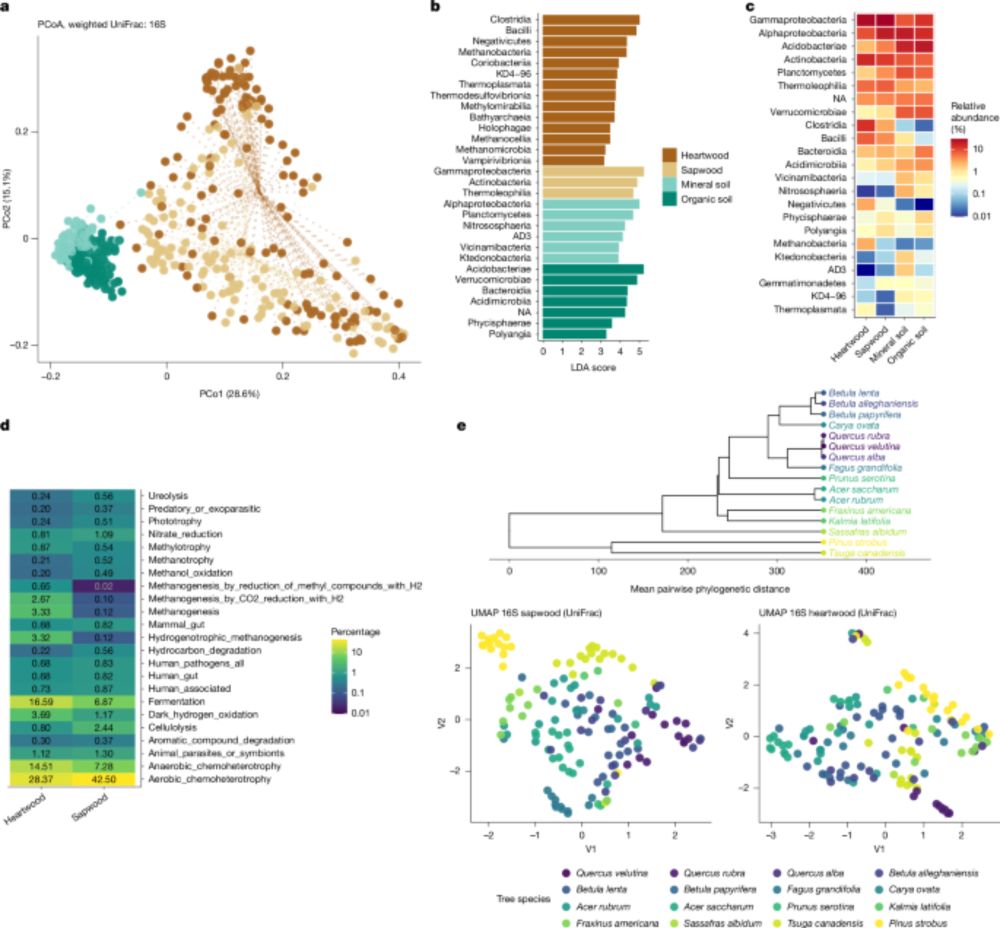
Hidden microbial world in trees🌳
Living wood hosts trillions of bacteria making trees a complex ecosystems with major roles in forest health and function.
#PlantMicro #MicroSky
www.nature.com/articles/s41...
academic.oup.com/bioinformati...

academic.oup.com/bioinformati...
#rnasky #microsky "cotranslational localization and translational accuracy, more than cotranslational protein folding, are major drivers of selective pressure on codon choice" in yeast here 💫
doi.org/10.1101/2025...

#rnasky #microsky "cotranslational localization and translational accuracy, more than cotranslational protein folding, are major drivers of selective pressure on codon choice" in yeast here 💫
doi.org/10.1101/2025...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵

Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵
github.com/lh3/longdust
github.com/lh3/longdust
My phage annotation tool, Phynteny, finally has a preprint and a brand new version powered by a cool AI transformer architecture and protein language models! #phagesky
www.biorxiv.org/content/10.1...

My phage annotation tool, Phynteny, finally has a preprint and a brand new version powered by a cool AI transformer architecture and protein language models! #phagesky
www.biorxiv.org/content/10.1...
🌐 afesm.foldseek.com
📄 www.biorxiv.org/content/10.1...

🌐 afesm.foldseek.com
📄 www.biorxiv.org/content/10.1...
You can get a sneak peek at the slides here: igor.martayan.org/slides-recom...
Come say hi if you'd like to chat, or just get one of these cute stickers!

You can get a sneak peek at the slides here: igor.martayan.org/slides-recom...
Come say hi if you'd like to chat, or just get one of these cute stickers!
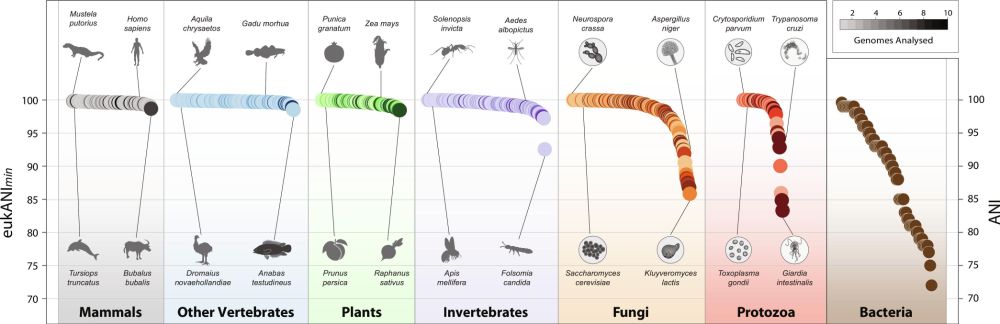
www.nature.com/articles/s41...
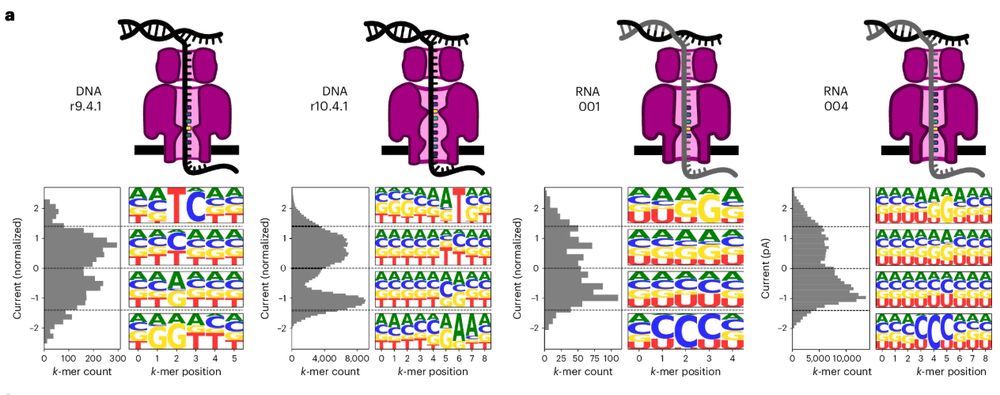
www.nature.com/articles/s41...
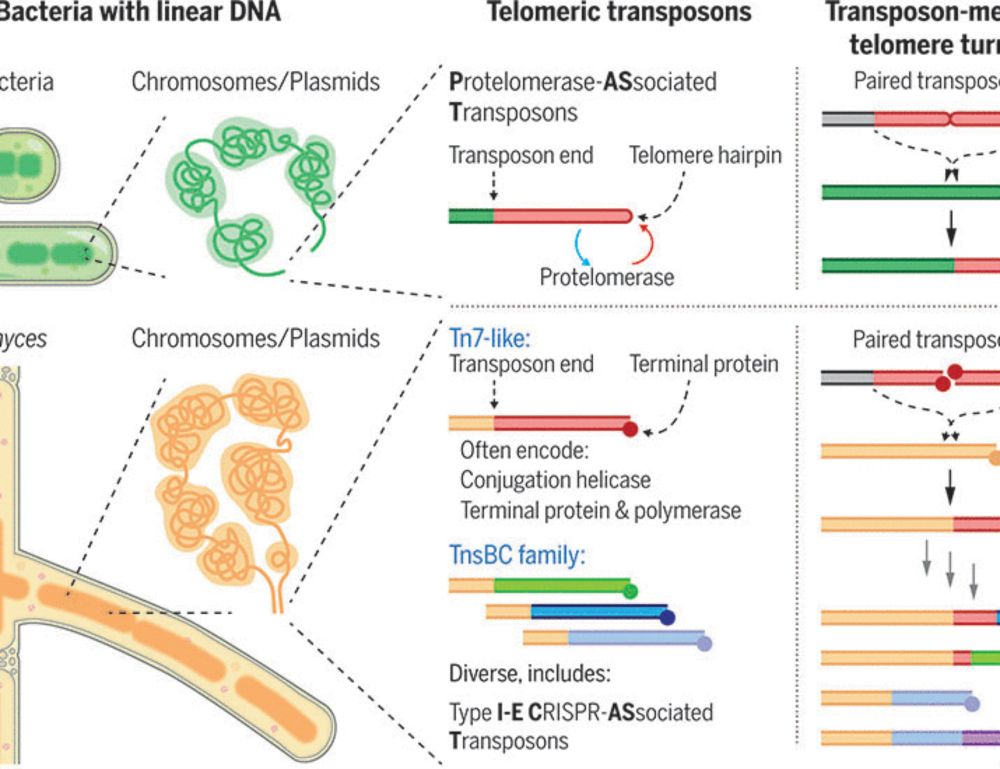
www.science.org/doi/10.1126/...
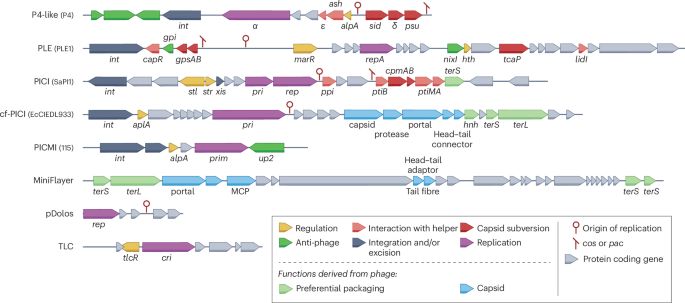
www.sciencedirect.com/science/arti...
www.sciencedirect.com/science/arti...
www.science.org/doi/10.1126/...
www.sciencedirect.com/science/arti...
www.sciencedirect.com/science/arti...
If so, you might this little project: github.com/rrwick/conda...
If so, you might this little project: github.com/rrwick/conda...