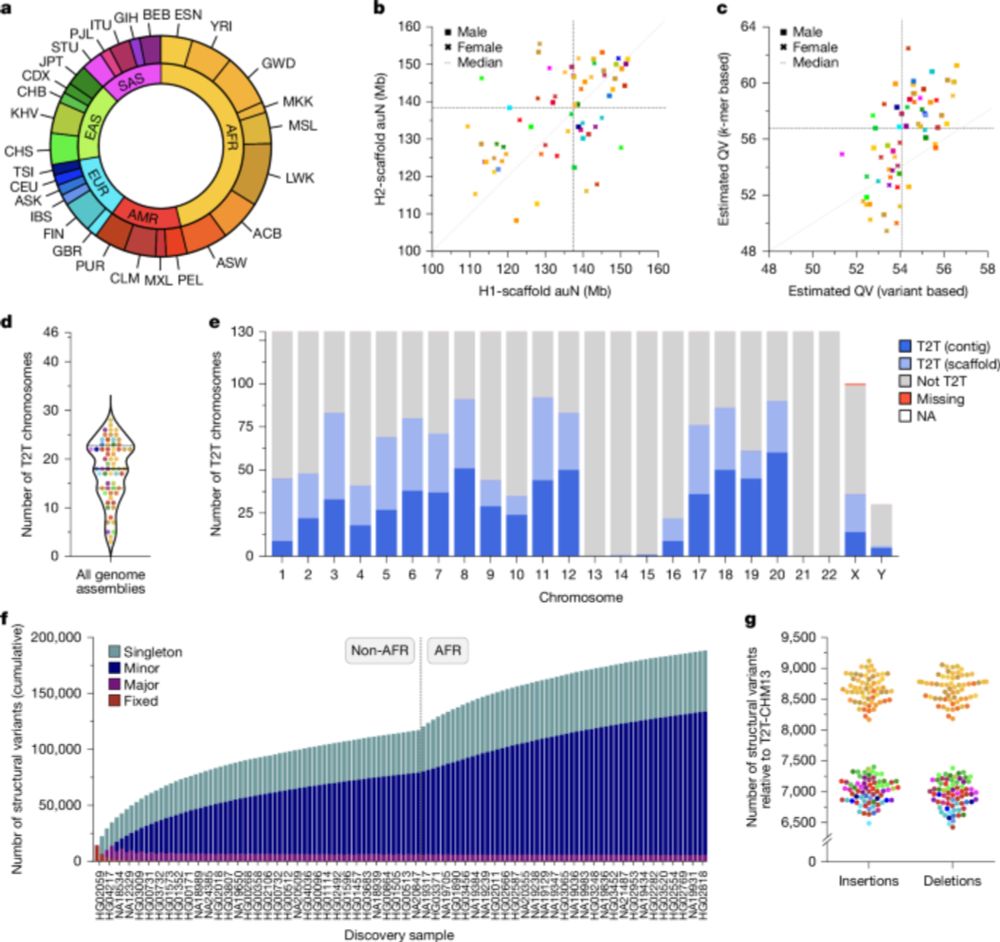
#epigenetics #centromere #TE
link.springer.com/collections/...
link.springer.com/collections/...
@natanaels.bsky.social and Anastasia Stolyarova, trying to think through what sets the mutation rate of a cell type in an animal species: www.biorxiv.org/content/10.6... 1/n

@natanaels.bsky.social and Anastasia Stolyarova, trying to think through what sets the mutation rate of a cell type in an animal species: www.biorxiv.org/content/10.6... 1/n

www.biorxiv.org/content/10.6...

Please spread the word; I think many people just outside my own circle could benefit from this :)
cc @rickbitloo.bsky.social
github.com/RagnarGrootK...
Please spread the word; I think many people just outside my own circle could benefit from this :)
cc @rickbitloo.bsky.social
github.com/RagnarGrootK...


Synergy between cis-regulatory elements can render cohesin dispensable for distal enhancer function
now revised and journal accepted at www.science.org/doi/10.1126/...
🧵👇
Synergy between cis-regulatory elements can render cohesin dispensable for distal enhancer function
now revised and journal accepted at www.science.org/doi/10.1126/...
🧵👇
Discover it in our new Nature paper! We show centromeres transition gradually via a mix of drift, selection, and sex, reaching new states that still work with the kinetochore.
👉 doi.org/10.1038/s41586-025-09779-1

Discover it in our new Nature paper! We show centromeres transition gradually via a mix of drift, selection, and sex, reaching new states that still work with the kinetochore.
👉 doi.org/10.1038/s41586-025-09779-1
www.nature.com/articles/s41...

www.nature.com/articles/s41...
doi.org/10.1016/j.ce...
We developed a new method called MCC ultra, which allows 3D chromatin structure to be visualised with a 1 base pair pixel size.

doi.org/10.1016/j.ce...
We developed a new method called MCC ultra, which allows 3D chromatin structure to be visualised with a 1 base pair pixel size.
Today, we report the discovery of telomerase homologs in a family of antiviral reverse transcriptases, revealing an unexpected evolutionary origin in bacteria.
doi.org/10.1101/2025...

Today, we report the discovery of telomerase homologs in a family of antiviral reverse transcriptases, revealing an unexpected evolutionary origin in bacteria.
doi.org/10.1101/2025...

Congrats Vincent Colot and team!
www.science.org/doi/10.1126/...

Congrats Vincent Colot and team!
www.science.org/doi/10.1126/...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
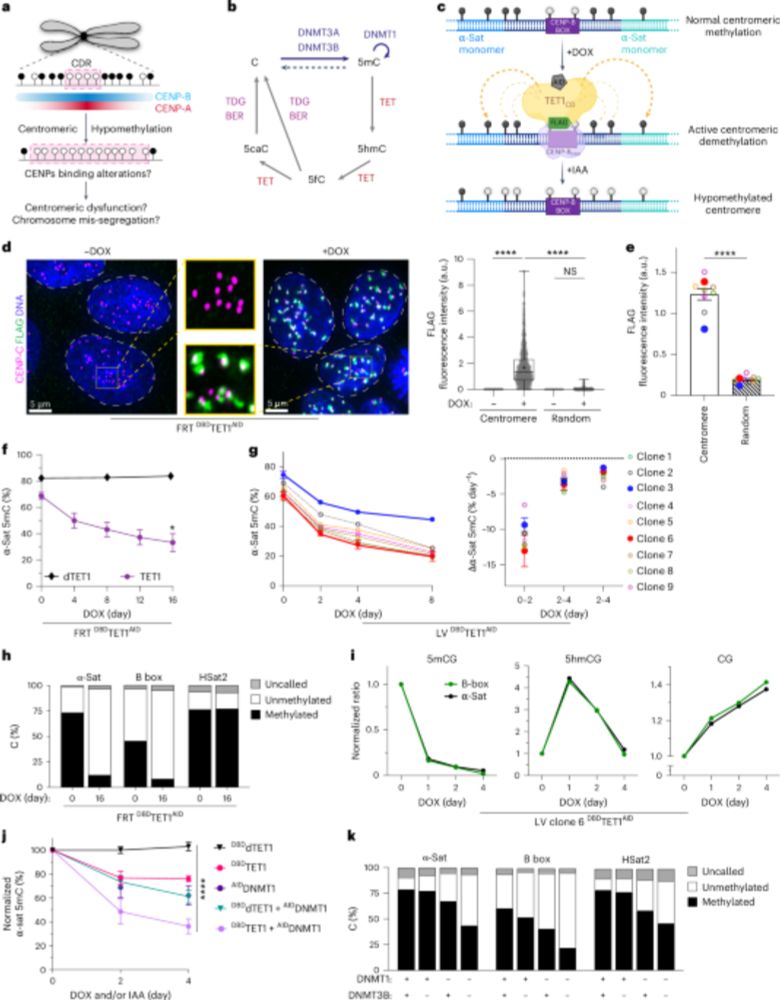

We present VICAR (VIral-induced Centromeric DNA Amplification and Recognition), a new defense system to detect viruses in the nucleus based on nuclear cGAS.
www.cell.com/cell/abstrac...
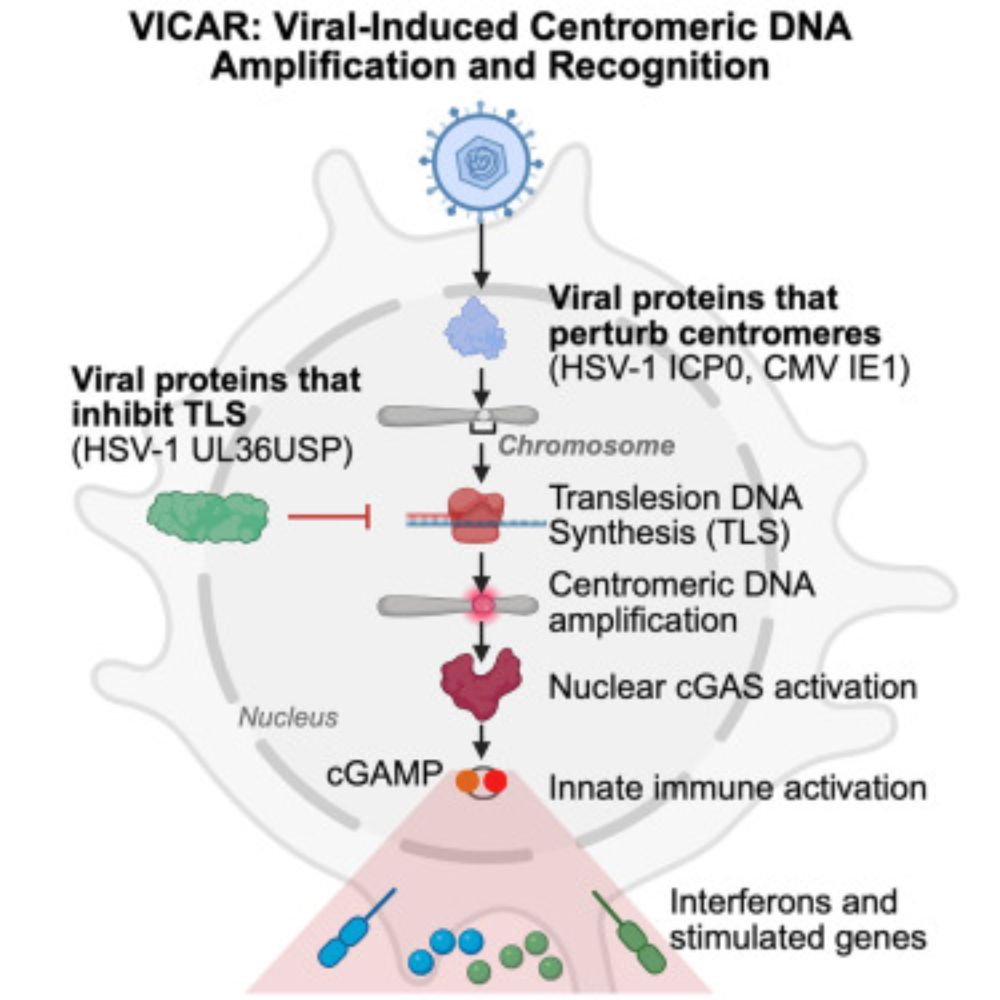
We present VICAR (VIral-induced Centromeric DNA Amplification and Recognition), a new defense system to detect viruses in the nucleus based on nuclear cGAS.
www.cell.com/cell/abstrac...