
Jan Mathony
@jmathony.bsky.social
Scientist with a passion for Synbio | Protein engineering | Optogenetics | ML
www.niopeklab.de/mathony-lab/
www.niopeklab.de/mathony-lab/
Reposted by Jan Mathony
With this, the last bit of my PhD at @embl.org is finally out!
We developed salad (sparse all-atom denoising), a family of blazing fast protein structure diffusion models.
Paper: nature.com/articles/s42256-…
Code: github.com/mjendrusch/salad
Data: zenodo.org/records/14711580
1/🧵
We developed salad (sparse all-atom denoising), a family of blazing fast protein structure diffusion models.
Paper: nature.com/articles/s42256-…
Code: github.com/mjendrusch/salad
Data: zenodo.org/records/14711580
1/🧵
‘Salad’ – a new AI model from EMBL scientists – offers major improvements in synthetic protein design.
Salad is significantly faster than comparable methods, and designing proteins that don't exist in nature can have applications in many scientific fields.
www.nature.com/articles/s42...
Salad is significantly faster than comparable methods, and designing proteins that don't exist in nature can have applications in many scientific fields.
www.nature.com/articles/s42...

September 24, 2025 at 12:27 PM
With this, the last bit of my PhD at @embl.org is finally out!
We developed salad (sparse all-atom denoising), a family of blazing fast protein structure diffusion models.
Paper: nature.com/articles/s42256-…
Code: github.com/mjendrusch/salad
Data: zenodo.org/records/14711580
1/🧵
We developed salad (sparse all-atom denoising), a family of blazing fast protein structure diffusion models.
Paper: nature.com/articles/s42256-…
Code: github.com/mjendrusch/salad
Data: zenodo.org/records/14711580
1/🧵
Reposted by Jan Mathony
Work by @bene837.bsky.social @jmathony.bsky.social @dominikniopek.bsky.social @uniheidelberg.bsky.social
ProDomino is a machine learning-based method that predicts domain insertion sites and helps guide the engineering of functional multi-domain proteins.
www.nature.com/articles/s41...
www.nature.com/articles/s41...
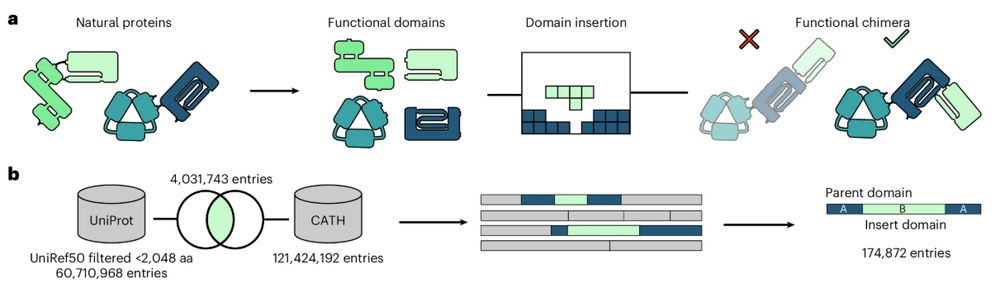
August 4, 2025 at 3:26 PM
Publication alert: Our paper on domain insertion predictions in proteins is now out in @natmethods.nature.com in its final form: rdcu.be/ey7w3
Also check out the nice perspective by @noahholzleitner.bsky.social and @grunewald.bsky.social : www.nature.com/articles/s41...
Also check out the nice perspective by @noahholzleitner.bsky.social and @grunewald.bsky.social : www.nature.com/articles/s41...

Rational engineering of allosteric protein switches by in silico prediction of domain insertion sites
Nature Methods - ProDomino is a machine leaning-based method, trained on a semisynthetic domain insertion dataset, to guide the engineering of protein domain recombination.
rdcu.be
August 4, 2025 at 12:28 PM
Publication alert: Our paper on domain insertion predictions in proteins is now out in @natmethods.nature.com in its final form: rdcu.be/ey7w3
Also check out the nice perspective by @noahholzleitner.bsky.social and @grunewald.bsky.social : www.nature.com/articles/s41...
Also check out the nice perspective by @noahholzleitner.bsky.social and @grunewald.bsky.social : www.nature.com/articles/s41...
Reposted by Jan Mathony
I'm very excited to finally share the main work of my PhD!
We explored the evolutionary dynamics of gene regulation and expression during gonad development in primates. We cover among others: X chromosome dynamics (incl. in a developing XXY testis), gene regulatory networks and cell type evolution.
We explored the evolutionary dynamics of gene regulation and expression during gonad development in primates. We cover among others: X chromosome dynamics (incl. in a developing XXY testis), gene regulatory networks and cell type evolution.
We are delighted to share our new preprint “The evolution of gene regulatory programs controlling gonadal development in primates” www.biorxiv.org/content/10.1...
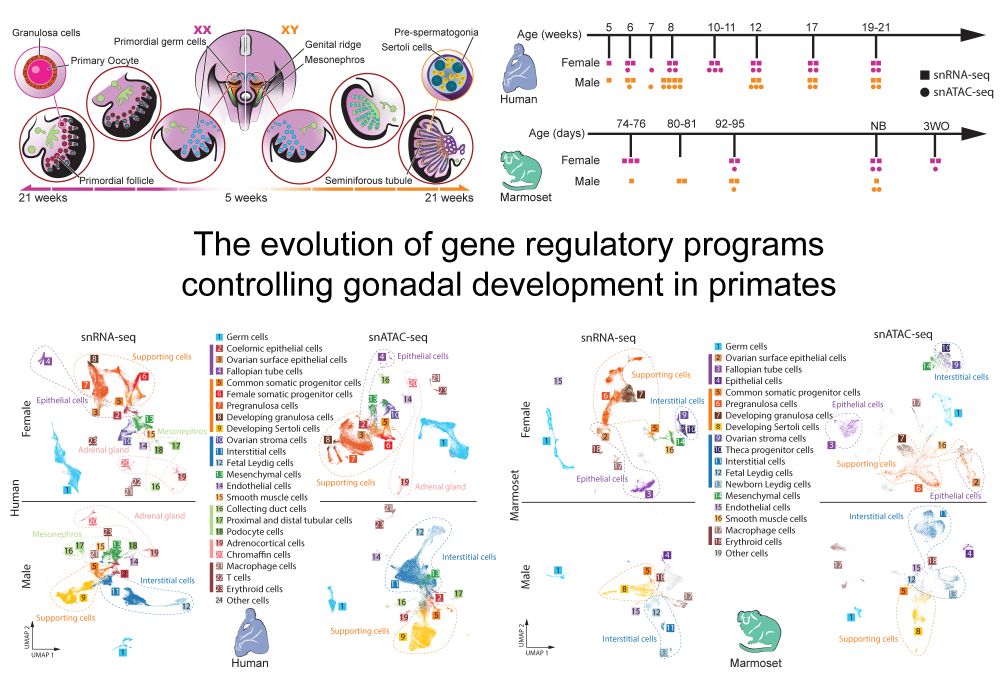
June 20, 2025 at 8:37 AM
I'm very excited to finally share the main work of my PhD!
We explored the evolutionary dynamics of gene regulation and expression during gonad development in primates. We cover among others: X chromosome dynamics (incl. in a developing XXY testis), gene regulatory networks and cell type evolution.
We explored the evolutionary dynamics of gene regulation and expression during gonad development in primates. We cover among others: X chromosome dynamics (incl. in a developing XXY testis), gene regulatory networks and cell type evolution.
Reposted by Jan Mathony
We are delighted to share our new preprint “The evolution of gene regulatory programs controlling gonadal development in primates” www.biorxiv.org/content/10.1...

June 20, 2025 at 8:13 AM
We are delighted to share our new preprint “The evolution of gene regulatory programs controlling gonadal development in primates” www.biorxiv.org/content/10.1...
Reposted by Jan Mathony
Happy to announce the first paper from my PhD at Korbel group at @embl.org has finally been published:
embopress.org/doi/full/10.1038…
Collaborating with @typaslab.bsky.social, @hennig-lab.bsky.social and the EMBL PEPCF, we designed de novo inhibitors to a bacterial phage defense system 1/🧵
embopress.org/doi/full/10.1038…
Collaborating with @typaslab.bsky.social, @hennig-lab.bsky.social and the EMBL PEPCF, we designed de novo inhibitors to a bacterial phage defense system 1/🧵
June 19, 2025 at 8:10 AM
Happy to announce the first paper from my PhD at Korbel group at @embl.org has finally been published:
embopress.org/doi/full/10.1038…
Collaborating with @typaslab.bsky.social, @hennig-lab.bsky.social and the EMBL PEPCF, we designed de novo inhibitors to a bacterial phage defense system 1/🧵
embopress.org/doi/full/10.1038…
Collaborating with @typaslab.bsky.social, @hennig-lab.bsky.social and the EMBL PEPCF, we designed de novo inhibitors to a bacterial phage defense system 1/🧵
Reposted by Jan Mathony
Inspired by how nature evolves trigger responsiveness through alternating pressures, we are excited to present POGO-PANCE and RAMPhaGE:
Phage-assisted evolution platforms for engineering allosteric protein switches under dynamic selection.
Preprint: doi.org/10.1101/2025...
Phage-assisted evolution platforms for engineering allosteric protein switches under dynamic selection.
Preprint: doi.org/10.1101/2025...

Phage-Assisted Evolution of Allosteric Protein Switches
Allostery, the transmission of locally induced conformational changes to distant functional sites, is a key mechanism for protein regulation. Artificial allosteric effectors enable remote manipulation...
doi.org
June 13, 2025 at 1:29 PM
Inspired by how nature evolves trigger responsiveness through alternating pressures, we are excited to present POGO-PANCE and RAMPhaGE:
Phage-assisted evolution platforms for engineering allosteric protein switches under dynamic selection.
Preprint: doi.org/10.1101/2025...
Phage-assisted evolution platforms for engineering allosteric protein switches under dynamic selection.
Preprint: doi.org/10.1101/2025...
New Preprint on phage-assisted evolution and retron-mediated mutagenesis for protein optimization.
Many congrats to @neuroscinikolai.bsky.social for this heroic effort and all other lab members involved!
Many congrats to @neuroscinikolai.bsky.social for this heroic effort and all other lab members involved!
Check out the new pre-print from our lab on phage-assisted evolution of light-switchable, allosteric proteins. Congrats to first author @neuroscinikolai.bsky.social, co-corresponding author @jmathony.bsky.social and everyone from the @niopeklab.bsky.social involved!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Phage-Assisted Evolution of Allosteric Protein Switches
Allostery, the transmission of locally induced conformational changes to distant functional sites, is a key mechanism for protein regulation. Artificial allosteric effectors enable remote manipulation...
www.biorxiv.org
June 13, 2025 at 11:03 AM
New Preprint on phage-assisted evolution and retron-mediated mutagenesis for protein optimization.
Many congrats to @neuroscinikolai.bsky.social for this heroic effort and all other lab members involved!
Many congrats to @neuroscinikolai.bsky.social for this heroic effort and all other lab members involved!
New paper alert! We introduce the modular allosteric thermo-control of protein activity. Employing the AsLOV2 domain and mutants thereof as thermoreceptors, we engineered diverse hybrid proteins, whose activity can be controlled by small temperature changes (37-40/41 °C).
doi.org/10.1101/2025...
doi.org/10.1101/2025...
doi.org
May 3, 2025 at 11:03 AM
New paper alert! We introduce the modular allosteric thermo-control of protein activity. Employing the AsLOV2 domain and mutants thereof as thermoreceptors, we engineered diverse hybrid proteins, whose activity can be controlled by small temperature changes (37-40/41 °C).
doi.org/10.1101/2025...
doi.org/10.1101/2025...
Reposted by Jan Mathony
Hey #RNA world, we are excited to share our RNA design engine #pyFuRNAce - an integrated tool for RNA origami experts and novices alike. Kudos to @monari-luca.bsky.social @floppleton.bsky.social et al! Preprint: www.biorxiv.org/content/10.1... Try it out at pyfurnace.de Feedback welcome!

PyFuRNAce: An integrated design engine for RNA origami
To realize the full potential of RNA nanotechnology and RNA origami, user-friendly design tools are needed. Here, we present pyFuRNAce, an open-source, Python-based software package with a graphical u...
www.biorxiv.org
April 20, 2025 at 12:29 PM
Hey #RNA world, we are excited to share our RNA design engine #pyFuRNAce - an integrated tool for RNA origami experts and novices alike. Kudos to @monari-luca.bsky.social @floppleton.bsky.social et al! Preprint: www.biorxiv.org/content/10.1... Try it out at pyfurnace.de Feedback welcome!
Reposted by Jan Mathony
Thank you to @yaseminsaplakoglu.bsky.social, who wrote a fantastic article for @quantamagazine.bsky.social about our studies on the evolution of the avian pallium. Love it! shorturl.at/vmTLq

Intelligence Evolved at Least Twice in Vertebrate Animals | Quanta Magazine
Complex neural circuits likely arose independently in birds and mammals, suggesting that vertebrates evolved intelligence multiple times.
shorturl.at
April 7, 2025 at 3:52 PM
Thank you to @yaseminsaplakoglu.bsky.social, who wrote a fantastic article for @quantamagazine.bsky.social about our studies on the evolution of the avian pallium. Love it! shorturl.at/vmTLq
We have an exciting new PhD opportunity!
If you are fsacinated by proteins and their dynamics and want to engineer them to develop new molecular tools: apply now!
We are a young and dynamic team combining state-of-the-art laboratory and bioinformatics approaches.
Please share.
If you are fsacinated by proteins and their dynamics and want to engineer them to develop new molecular tools: apply now!
We are a young and dynamic team combining state-of-the-art laboratory and bioinformatics approaches.
Please share.

April 1, 2025 at 11:52 AM
We have an exciting new PhD opportunity!
If you are fsacinated by proteins and their dynamics and want to engineer them to develop new molecular tools: apply now!
We are a young and dynamic team combining state-of-the-art laboratory and bioinformatics approaches.
Please share.
If you are fsacinated by proteins and their dynamics and want to engineer them to develop new molecular tools: apply now!
We are a young and dynamic team combining state-of-the-art laboratory and bioinformatics approaches.
Please share.
Reposted by Jan Mathony
What a week! I defended my PhD on Monday, and now my first first-author paper was published in @science.org.
shorturl.at/fvIGZ
I am so incredibly grateful to everyone who made this possible! Especially @kaessmannlab.bsky.social and the García-Moreno lab 💛
shorturl.at/fvIGZ
I am so incredibly grateful to everyone who made this possible! Especially @kaessmannlab.bsky.social and the García-Moreno lab 💛


February 14, 2025 at 6:27 PM
What a week! I defended my PhD on Monday, and now my first first-author paper was published in @science.org.
shorturl.at/fvIGZ
I am so incredibly grateful to everyone who made this possible! Especially @kaessmannlab.bsky.social and the García-Moreno lab 💛
shorturl.at/fvIGZ
I am so incredibly grateful to everyone who made this possible! Especially @kaessmannlab.bsky.social and the García-Moreno lab 💛
Reposted by Jan Mathony
So excited to announce that our study on the development and evolution of pallial cell types and structures in birds, led by @bassi-z.bsky.social, is now out in @science.org!
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...

Developmental origins and evolution of pallial cell types and structures in birds
Innovations in the pallium likely facilitated the evolution of advanced cognitive abilities in birds. We therefore scrutinized its cellular composition and evolution using cell type atlases from chick...
www.science.org
February 13, 2025 at 10:17 PM
So excited to announce that our study on the development and evolution of pallial cell types and structures in birds, led by @bassi-z.bsky.social, is now out in @science.org!
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
Reposted by Jan Mathony
Excited to announce our optogenetic transcriptional deactivation toolbox is now out in its final form at Nucleic Acids research: academic.oup.com/nar/advance-....

A modular toolbox for the optogenetic deactivation of transcription
Abstract. Light-controlled transcriptional activation is a commonly used optogenetic strategy that allows researchers to regulate gene expression with high
academic.oup.com
December 16, 2024 at 12:54 PM
Excited to announce our optogenetic transcriptional deactivation toolbox is now out in its final form at Nucleic Acids research: academic.oup.com/nar/advance-....
I am super excited and grateful to be funded by the BW-Stiftung within the Postdoc Elite Program. Looking forward to bringing exciting new research from the drawing board into the lab. Many thanks to all my amazing colleagues for all the continued collaboration and support!
December 9, 2024 at 12:20 PM
I am super excited and grateful to be funded by the BW-Stiftung within the Postdoc Elite Program. Looking forward to bringing exciting new research from the drawing board into the lab. Many thanks to all my amazing colleagues for all the continued collaboration and support!
Reposted by Jan Mathony
We updated our BindCraft preprint with lots of new exciting results! We release all our binder sequences and models, include more in silico analysis, novel design targets, and present AAV retargeting to specific cell types using de novo binders!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
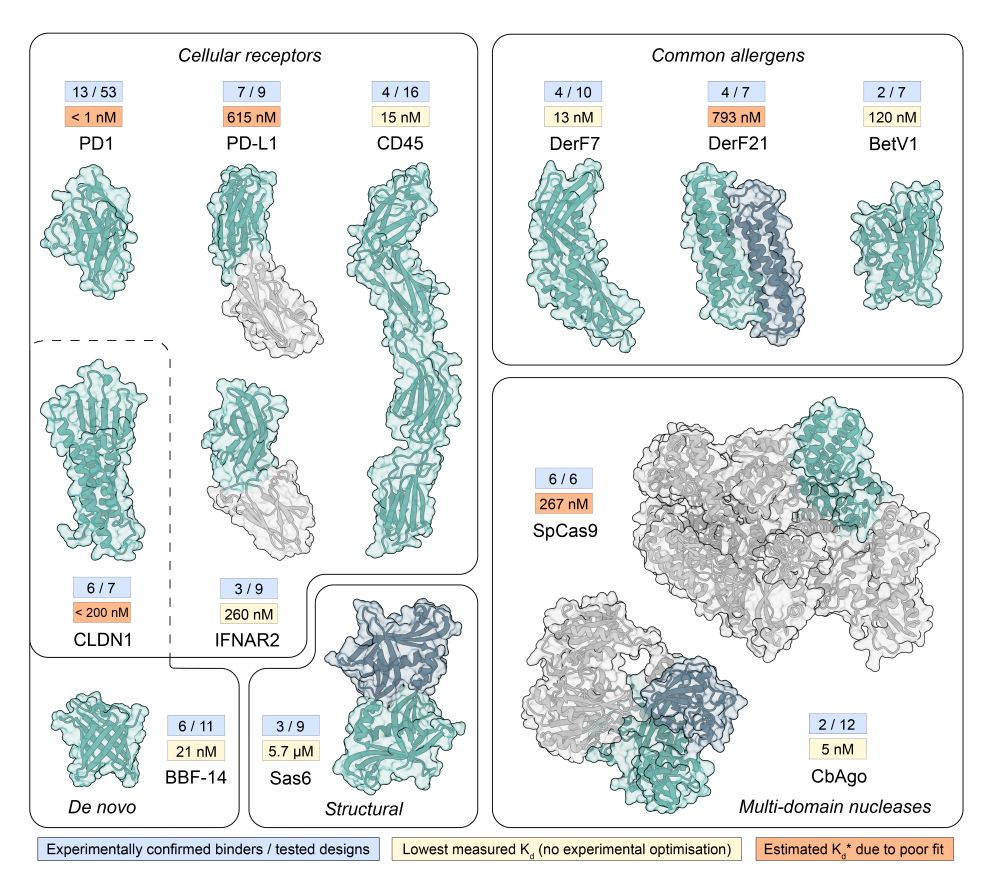
December 8, 2024 at 8:27 AM
We updated our BindCraft preprint with lots of new exciting results! We release all our binder sequences and models, include more in silico analysis, novel design targets, and present AAV retargeting to specific cell types using de novo binders!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Interested in allosteric protein switches?
Check our ProDomino - our new model for the prediction of domain insertion sites. With ProDomino we engineered several potent inducible proteins including Cas9 and Cas12a variants.
Check our ProDomino - our new model for the prediction of domain insertion sites. With ProDomino we engineered several potent inducible proteins including Cas9 and Cas12a variants.
We are thrilled to share ProDomino a model for the prediction of domain insertion sites in proteins. Our approach enables the simple and rapid engineering of highly potent switchable proteins, as we exemplify by creating novel inducible variants of Cas9 and Cas12a.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Rational engineering of allosteric protein switches by in silico prediction of domain insertion sites
Domain insertion engineering is a powerful approach to juxtapose otherwise separate biological functions, resulting in proteins with new-to-nature activities. A prominent example are switchable protei...
www.biorxiv.org
December 5, 2024 at 10:04 AM
Interested in allosteric protein switches?
Check our ProDomino - our new model for the prediction of domain insertion sites. With ProDomino we engineered several potent inducible proteins including Cas9 and Cas12a variants.
Check our ProDomino - our new model for the prediction of domain insertion sites. With ProDomino we engineered several potent inducible proteins including Cas9 and Cas12a variants.
Multi-modal control of gene editing with inducible anti-CRISPR proteins!
Congrats to first authors @lucabrenker.bsky.social, Sabine and Felix.
Congrats to first authors @lucabrenker.bsky.social, Sabine and Felix.
New pre-print from our group reporting engineered, broad-spectrum anti-CRISPR proteins based on AcrIIA5, a type II inhibitor, and AcrVA1, a type V inhibitor, for opto- and chemogenetic control of CRISPR-Cas9 and -Cas12a:
www.biorxiv.org/content/10.1...
(1/3)
www.biorxiv.org/content/10.1...
(1/3)

A Versatile Anti-CRISPR Platform for Opto- and Chemogenetic Control of CRISPR-Cas9 and Cas12 across a Wide Range of Orthologs
CRISPR-Cas technologies have revolutionized life sciences by enabling programmable genome editing across diverse organisms. Achieving dynamic and precise control over CRISPR-Cas activity with exogenou...
www.biorxiv.org
November 26, 2024 at 1:04 PM
Multi-modal control of gene editing with inducible anti-CRISPR proteins!
Congrats to first authors @lucabrenker.bsky.social, Sabine and Felix.
Congrats to first authors @lucabrenker.bsky.social, Sabine and Felix.
Reposted by Jan Mathony
Now out in Nucleic Acids Research: A deep mutational scanning platform to characterize the fitness landscape of anti-CRISPR proteins: doi.org/10.1093/nar/...
(1/4)
(1/4)

A deep mutational scanning platform to characterize the fitness landscape of anti-CRISPR proteins
Abstract. Deep mutational scanning is a powerful method for exploring the mutational fitness landscape of proteins. Its adaptation to anti-CRISPR proteins,
doi.org
November 26, 2024 at 11:30 AM
Now out in Nucleic Acids Research: A deep mutational scanning platform to characterize the fitness landscape of anti-CRISPR proteins: doi.org/10.1093/nar/...
(1/4)
(1/4)
Reposted by Jan Mathony
New pre-print from our group reporting engineered, broad-spectrum anti-CRISPR proteins based on AcrIIA5, a type II inhibitor, and AcrVA1, a type V inhibitor, for opto- and chemogenetic control of CRISPR-Cas9 and -Cas12a:
www.biorxiv.org/content/10.1...
(1/3)
www.biorxiv.org/content/10.1...
(1/3)

A Versatile Anti-CRISPR Platform for Opto- and Chemogenetic Control of CRISPR-Cas9 and Cas12 across a Wide Range of Orthologs
CRISPR-Cas technologies have revolutionized life sciences by enabling programmable genome editing across diverse organisms. Achieving dynamic and precise control over CRISPR-Cas activity with exogenou...
www.biorxiv.org
November 26, 2024 at 11:34 AM
New pre-print from our group reporting engineered, broad-spectrum anti-CRISPR proteins based on AcrIIA5, a type II inhibitor, and AcrVA1, a type V inhibitor, for opto- and chemogenetic control of CRISPR-Cas9 and -Cas12a:
www.biorxiv.org/content/10.1...
(1/3)
www.biorxiv.org/content/10.1...
(1/3)
Reposted by Jan Mathony
A Versatile Anti-CRISPR Platform for Opto- and Chemogenetic Control of CRISPR-Cas9 and Cas12 across a Wide Range of Orthologs https://www.biorxiv.org/content/10.1101/2024.11.25.625186v1

A Versatile Anti-CRISPR Platform for Opto- and Chemogenetic Control of CRISPR-Cas9 and Cas12 across a Wide Range of Orthologs https://www.biorxiv.org/content/10.1101/2024.11.25.625186v1
CRISPR-Cas technologies have revolutionized life sciences by enabling programmable genome editing ac
www.biorxiv.org
November 26, 2024 at 5:04 AM
A Versatile Anti-CRISPR Platform for Opto- and Chemogenetic Control of CRISPR-Cas9 and Cas12 across a Wide Range of Orthologs https://www.biorxiv.org/content/10.1101/2024.11.25.625186v1
Our paper on DMS of anti-CRISPR proteins is out in it's final form Nucleic Acids Research!
Many congrats to first authors Tobias and Michael! It was lots of fun to see the story develop.
academic.oup.com/nar/advance-...
Many congrats to first authors Tobias and Michael! It was lots of fun to see the story develop.
academic.oup.com/nar/advance-...

A deep mutational scanning platform to characterize the fitness landscape of anti-CRISPR proteins
Abstract. Deep mutational scanning is a powerful method for exploring the mutational fitness landscape of proteins. Its adaptation to anti-CRISPR proteins,
academic.oup.com
November 22, 2024 at 6:19 PM
Our paper on DMS of anti-CRISPR proteins is out in it's final form Nucleic Acids Research!
Many congrats to first authors Tobias and Michael! It was lots of fun to see the story develop.
academic.oup.com/nar/advance-...
Many congrats to first authors Tobias and Michael! It was lots of fun to see the story develop.
academic.oup.com/nar/advance-...

