ML/Bio, genomic language models, epigenetics
Our work -- led by Sanjit Batra and Alan Cabrera when they were in @yun-s-song.bsky.social ’s and Isaac Hilton’s labs -- tries to answer this.
🧵🧬🧪
elifesciences.org/reviewed-pre...
Our work -- led by Sanjit Batra and Alan Cabrera when they were in @yun-s-song.bsky.social ’s and Isaac Hilton’s labs -- tries to answer this.
🧵🧬🧪
elifesciences.org/reviewed-pre...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
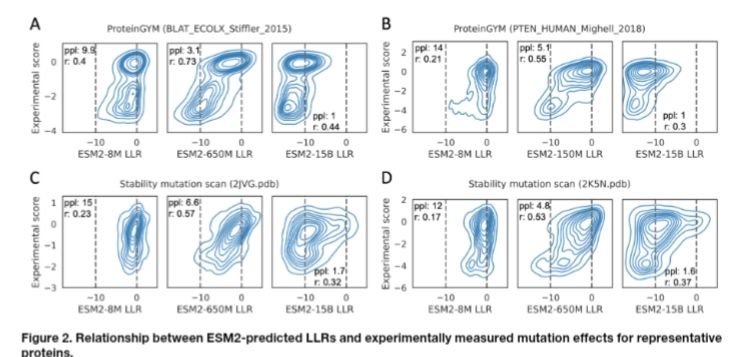
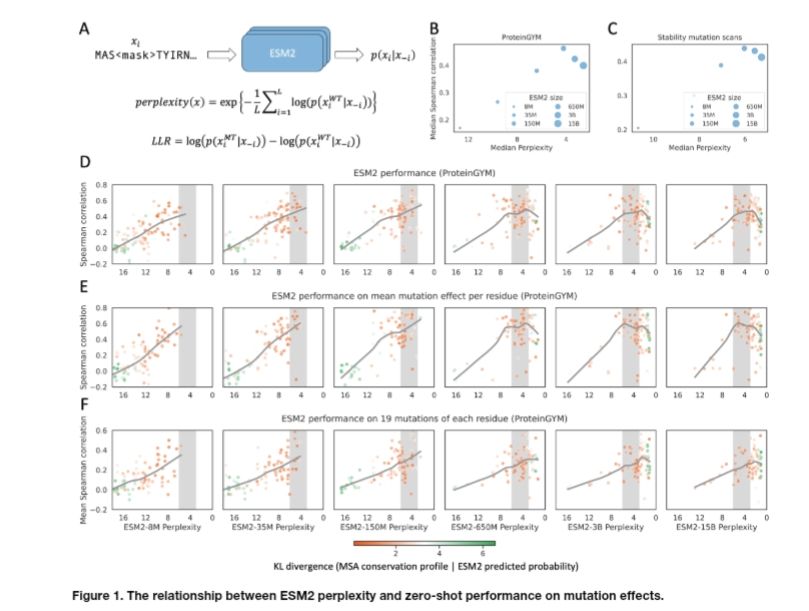
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
A 🧵about what it can do:
#SynBio #DeepLearning #GeneRegulation

www.biorxiv.org/content/10.1...
A 🧵about what it can do:
#SynBio #DeepLearning #GeneRegulation
But then I found the paper "Mechanistic?" by
@nsaphra.bsky.social and @sarah-nlp.bsky.social, which clarified things.

But then I found the paper "Mechanistic?" by
@nsaphra.bsky.social and @sarah-nlp.bsky.social, which clarified things.
arxiv.org/abs/2411.11158

arxiv.org/abs/2411.11158

