@annarogers.bsky.social on why not to rely on LLM AI to write for you in graduate school. hackingsemantics.xyz/2024/writing/

@annarogers.bsky.social on why not to rely on LLM AI to write for you in graduate school. hackingsemantics.xyz/2024/writing/
www.nature.com/articles/s41...
@pamsharmamdphd.bsky.social
www.nature.com/articles/s41...
@pamsharmamdphd.bsky.social
www.biorxiv.org/content/10.1...


www.biorxiv.org/content/10.1...
Kuang X, et al. (2024) SuperWater: Predicting water molecule positions on protein structures by generative AI. doi.org/10.1101/2024...
a generative model using score-based diffusion models & equivariant NNs, heavy GPU required for memory-intensive reverse diffusion steps
github.com/kuangxh9/Sup...

Kuang X, et al. (2024) SuperWater: Predicting water molecule positions on protein structures by generative AI. doi.org/10.1101/2024...
a generative model using score-based diffusion models & equivariant NNs, heavy GPU required for memory-intensive reverse diffusion steps
github.com/kuangxh9/Sup...
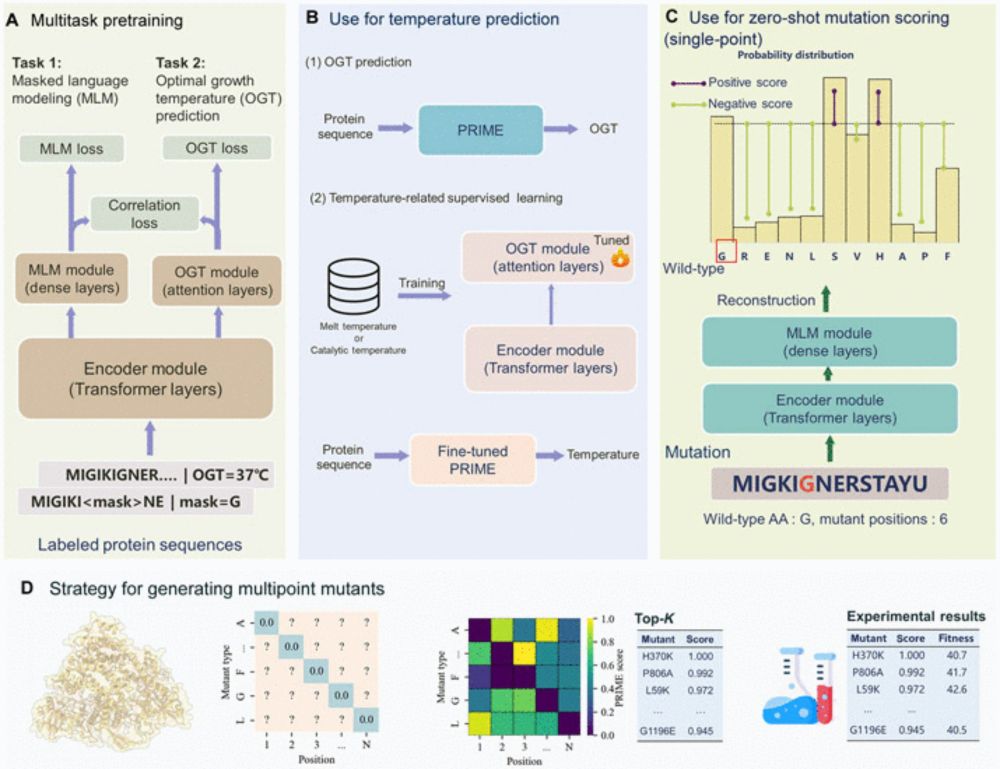
Publication of 10 new foundation models of Proteins, DNA, RNA, methylation, cells, and interactions, evolution, and design in the past couple of weeks!
Unprecedented progress, reviewed in the new Ground Truths
erictopol.substack.com/p/learning-t...

Publication of 10 new foundation models of Proteins, DNA, RNA, methylation, cells, and interactions, evolution, and design in the past couple of weeks!
Unprecedented progress, reviewed in the new Ground Truths
erictopol.substack.com/p/learning-t...

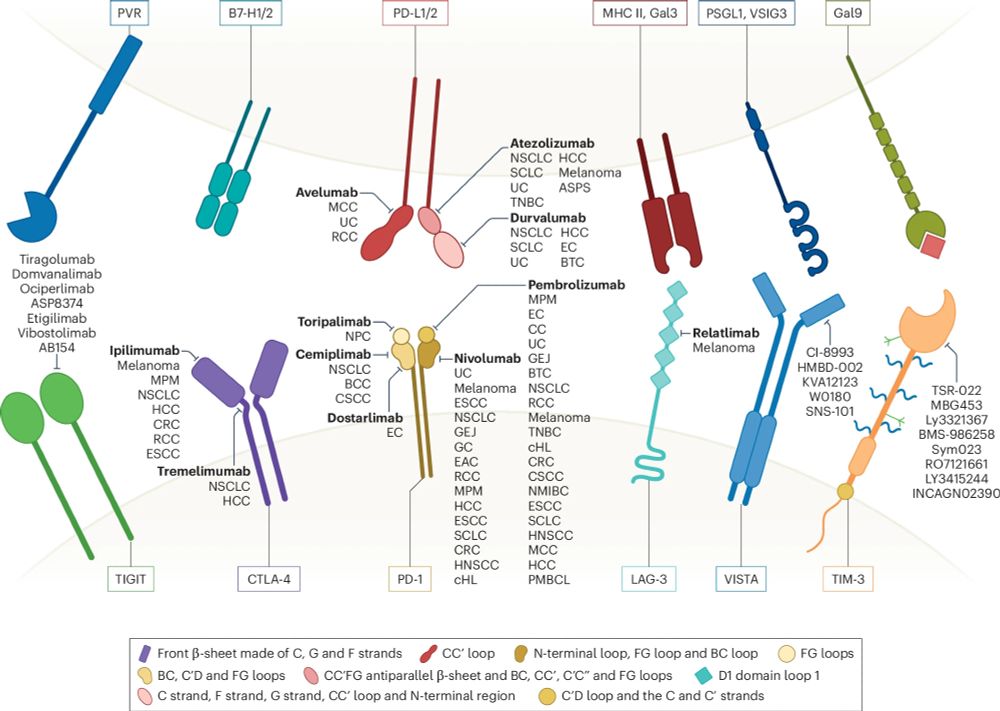
www.nature.com/articles/s41...
nabla-public.s3.us-east-1.amazonaws.com/2024_Nabla_J...

www.nature.com/articles/s41...
nabla-public.s3.us-east-1.amazonaws.com/2024_Nabla_J...



Here’s a link to the manuscript in NAR.



pubs.acs.org/doi/10.1021/...
@pabloarantes.bsky.social & team also made it into a Colab notebook:
colab.research.google.com/drive/1GGJKr...
Give it a try & send feedback!

pubs.acs.org/doi/10.1021/...
@pabloarantes.bsky.social & team also made it into a Colab notebook:
colab.research.google.com/drive/1GGJKr...
Give it a try & send feedback!
www.biorxiv.org/content/10.1...
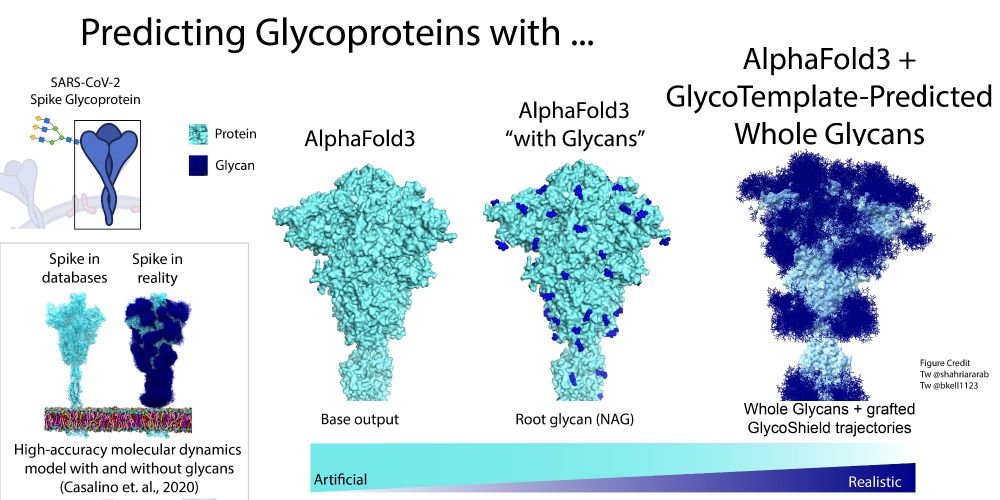
www.biorxiv.org/content/10.1...
📄 www.biorxiv.org/content/10.1...
💾 mmseqs.com
🗞️ developer.nvidia.com/blog/boost-a...

📄 www.biorxiv.org/content/10.1...
💾 mmseqs.com
🗞️ developer.nvidia.com/blog/boost-a...