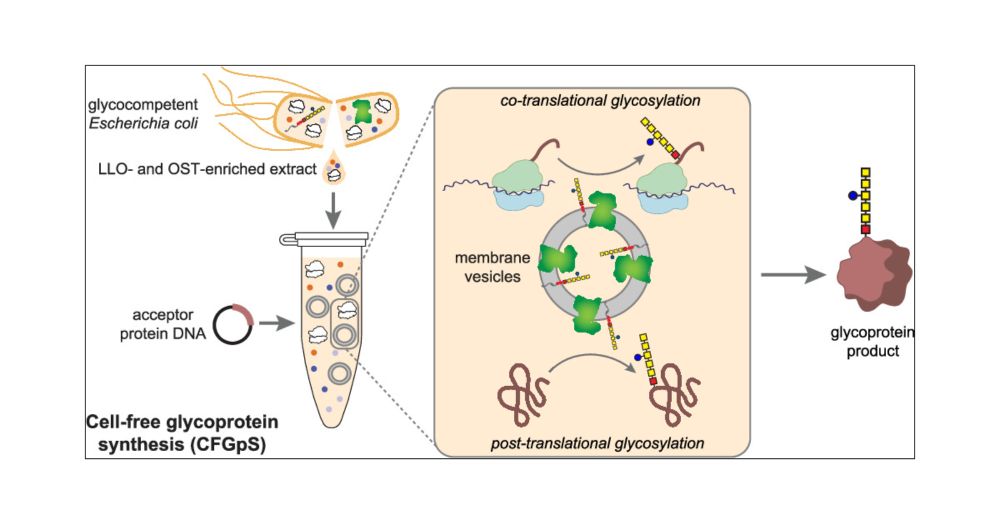

Hi all! We’re hiring at our brand new Center for Glycocalyx Research, across all areas - but I am also looking for postdocs to join my group specifically!
Hi all! We’re hiring at our brand new Center for Glycocalyx Research, across all areas - but I am also looking for postdocs to join my group specifically!

Hi all! We’re hiring at our brand new Center for Glycocalyx Research, across all areas - but I am also looking for postdocs to join my group specifically!

Our latest in @natrevneuro.nature.com
Link: rdcu.be/eMX3E
@jeffcolgren.bsky.social @msarscentre.bsky.social

Our latest in @natrevneuro.nature.com
Link: rdcu.be/eMX3E
@jeffcolgren.bsky.social @msarscentre.bsky.social


www.kearnslab.org


www.kearnslab.org
pubs.acs.org/doi/10.1021/...
github.com/snijderlab/a...
github.com/jspaezp/rust...

Hi all! We’re hiring at our brand new Center for Glycocalyx Research, across all areas - but I am also looking for postdocs to join my group specifically!

Hi all! We’re hiring at our brand new Center for Glycocalyx Research, across all areas - but I am also looking for postdocs to join my group specifically!

Hi all! We’re hiring at our brand new Center for Glycocalyx Research, across all areas - but I am also looking for postdocs to join my group specifically!


www.nature.com/articles/s41...

www.nature.com/articles/s41...