

pubs.acs.org/doi/10.1021/...
New paper from our group in Analytical Chemistry. Anne Sanner with support from Robert Hardt devised an approach to combine DIA analyses and scheduled PRM assays,
weiterlesen

weiterlesen
---
#proteomics #prot-other

---
#proteomics #prot-other
https://www.stocktitan.net/news/SEER/seer-s-proteograph-platform-enables-unprecedented-20-000-sample-j5bux0zunuwx.html

https://www.stocktitan.net/news/SEER/seer-s-proteograph-platform-enables-unprecedented-20-000-sample-j5bux0zunuwx.html
---
#proteomics #prot-paper
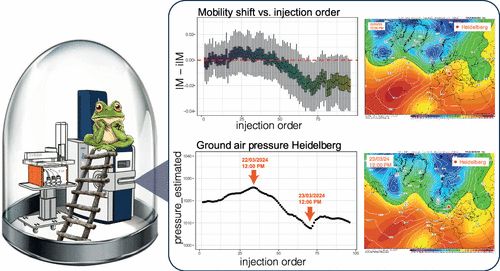
---
#proteomics #prot-paper
Our goal was to share a vision of #OSS #proteomics for us to build toward, and propose some ways to get there 🚀
I’m blown away by how many folks contributed and how much it evolved beyond just my voice 🙌
pubs.acs.org/doi/10.1021/...
New paper from our group in Analytical Chemistry. Anne Sanner with support from Robert Hardt devised an approach to combine DIA analyses and scheduled PRM assays,

pubs.acs.org/doi/10.1021/...
New paper from our group in Analytical Chemistry. Anne Sanner with support from Robert Hardt devised an approach to combine DIA analyses and scheduled PRM assays,
Most love the marketing. 5,000 proteins? 10,000 proteins? OLink was used in UK Biobank? Wow, mass spec is sooooo last year. Except life is not that simple.
I know why they do this but in the long run as it casts doubt on affinity approaches as a whole.
We will think lots of data is generated that is trash and won’t know which
Most love the marketing. 5,000 proteins? 10,000 proteins? OLink was used in UK Biobank? Wow, mass spec is sooooo last year. Except life is not that simple.

Also, next week see Lindsay et al. battle countless quant foes (not each other!) in the second US HUPO LFQ Battle Royale.

Also, next week see Lindsay et al. battle countless quant foes (not each other!) in the second US HUPO LFQ Battle Royale.



