As we all do, 45k citations and counting! 👏🏻👏🏻👏🏻
news.yale.edu/2025/11/18/d...

As we all do, 45k citations and counting! 👏🏻👏🏻👏🏻
news.yale.edu/2025/11/18/d...

Joana Bustamante's first first-author preprint in our group and us developing a model for thermal conductivity!
Feedback welcome!
arxiv.org/abs/2510.23133
#compchem

Joana Bustamante's first first-author preprint in our group and us developing a model for thermal conductivity!
Feedback welcome!
arxiv.org/abs/2510.23133
#compchem
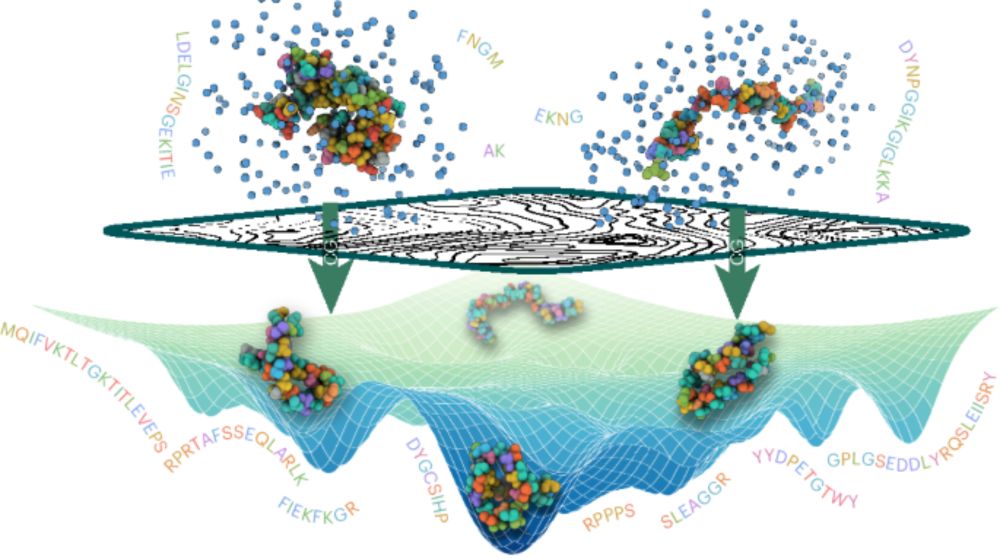
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...

See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
Time-dependent and -resolved experiments combined with computation provide a view on molecular dynamics beyond that available from static, ensemble-averaged experiments
Review w @dariagusew.bsky.social & Carl G Henning Hansen
doi.org/10.48550/arX...

Time-dependent and -resolved experiments combined with computation provide a view on molecular dynamics beyond that available from static, ensemble-averaged experiments
Review w @dariagusew.bsky.social & Carl G Henning Hansen
doi.org/10.48550/arX...
Interfolio link: apply.interfolio.com/174360
PLEASE, share widely across the blue skies!
Let me briefly explain what we're looking for:
1/10

Interfolio link: apply.interfolio.com/174360
PLEASE, share widely across the blue skies!
Let me briefly explain what we're looking for:
1/10

people.idsia.ch/~juergen/who...

people.idsia.ch/~juergen/who...
Please find our preprint here.
doi.org/10.26434/che...
#compchemsky

Please find our preprint here.
doi.org/10.26434/che...
#compchemsky
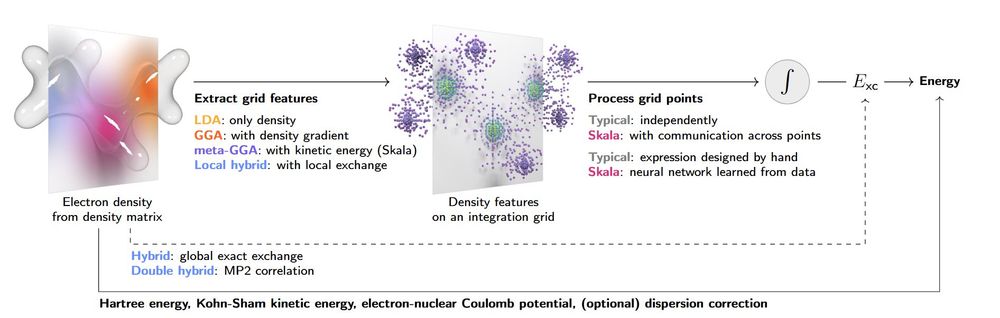
I am so proud of my group for this work! Particularly first authors Nick Charron, Klara Bonneau, Aldo Pasos-Trejo, Andrea Guljas.
I am so proud of my group for this work! Particularly first authors Nick Charron, Klara Bonneau, Aldo Pasos-Trejo, Andrea Guljas.
Workflows for computational materials science that are ready to be used!!!
#compchem
Workflows for computational materials science that are ready to be used!!!
#compchem
In a fantastic teamwork, @mcagiada.bsky.social and @emilthomasen.bsky.social developed AF2χ to generate conformational ensembles representing side-chain dynamics using AF2 💃
Code: github.com/KULL-Centre/...
Colab: github.com/matteo-cagia...
In a fantastic teamwork, @mcagiada.bsky.social and @emilthomasen.bsky.social developed AF2χ to generate conformational ensembles representing side-chain dynamics using AF2 💃
Code: github.com/KULL-Centre/...
Colab: github.com/matteo-cagia...
Just out in Digital Discovery:
doi.org/10.1039/D4DD...
#compchem

Just out in Digital Discovery:
doi.org/10.1039/D4DD...
#compchem
Nearly all teaching material including videos of our atomate2 school is already or will be online:
www.cecam.org/workshop-det...
#compchem
@virtualatoms.bsky.social @naikaakash.bsky.social and many more not on here 😀
Nearly all teaching material including videos of our atomate2 school is already or will be online:
www.cecam.org/workshop-det...
#compchem
@virtualatoms.bsky.social @naikaakash.bsky.social and many more not on here 😀
40 Spiele
29 Siege
5 Remis
6 Niederlagen
110:38 Tore
#B04FCB
>> Und SECHS Halbzeiten gg Leverkusen ohne Gegentor.
Github: github.com/BAMeScience/...
Paper: www.nature.com/articles/s41...
Many thanks to everyone involved 🙌 #MachineLearning #MassSpec #Metabolomics #FIORA
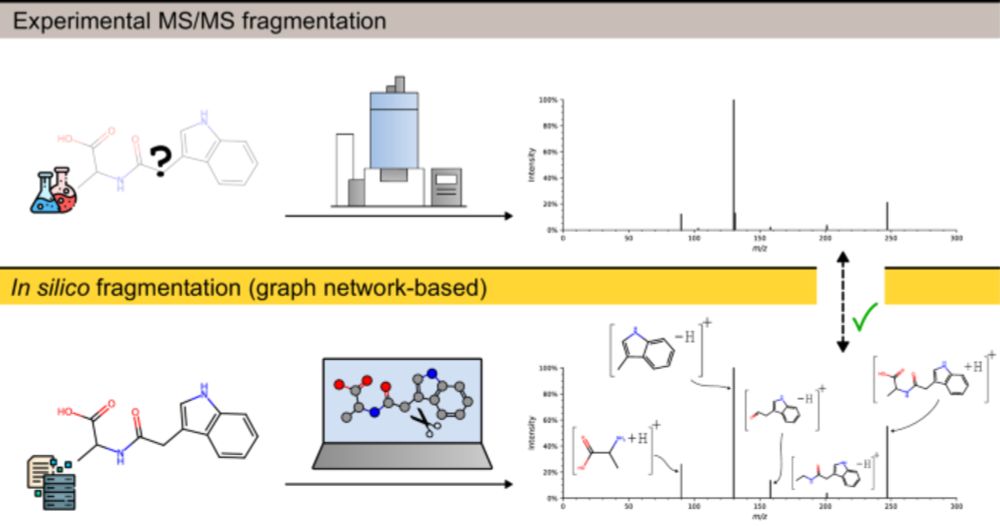
Github: github.com/BAMeScience/...
Paper: www.nature.com/articles/s41...
Many thanks to everyone involved 🙌 #MachineLearning #MassSpec #Metabolomics #FIORA
"Velocity Jumps for Molecular Dynamics"
pubs.acs.org/doi/10.1021/...
We introduce the Velocity Jumps approach, denoted as JUMP, a new class of Molecular dynamics integrators, replacing the Langevin dynamics. Amazing work by Nicolai Gouraud. #compchemsky

"Velocity Jumps for Molecular Dynamics"
pubs.acs.org/doi/10.1021/...
We introduce the Velocity Jumps approach, denoted as JUMP, a new class of Molecular dynamics integrators, replacing the Langevin dynamics. Amazing work by Nicolai Gouraud. #compchemsky
We are looking for candidates with a strong background in molecular dynamics simulations of membrane protein interactions to unravel the role of lipids in CD95 oligomerization and signaling!
Please apply by March 28!
#LipidTime @bzh-hd.bsky.social
˚⋆🔬Lipid-Mediated Regulation of CD95 #Oligomerization and Signaling🧪🥽🧠⋆˚
Applications on the Alliance website:
www.health-life-sciences.de/opportunitie...
@dkfz.bsky.social
@fabiololicato.com @bzh-hd.bsky.social
@hlsalliance.bsky.social
#LipidTime #ProteinDesign
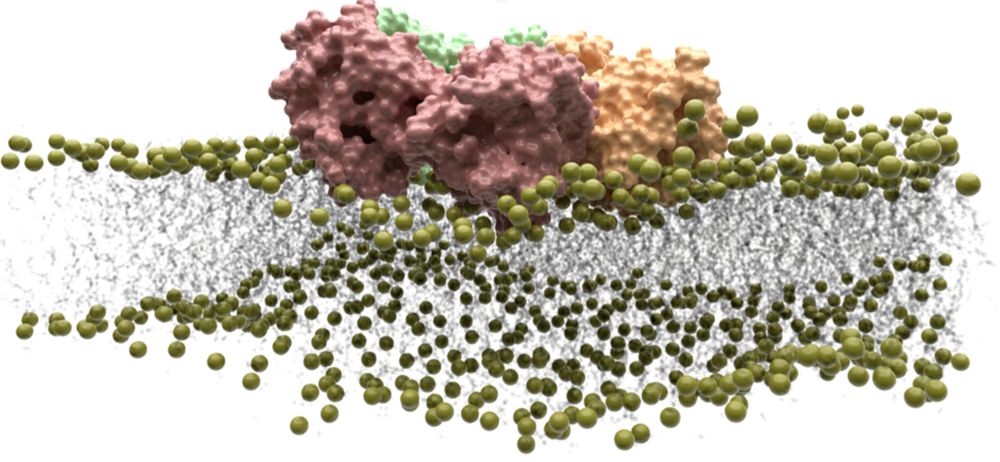
We are looking for candidates with a strong background in molecular dynamics simulations of membrane protein interactions to unravel the role of lipids in CD95 oligomerization and signaling!
Please apply by March 28!
#LipidTime @bzh-hd.bsky.social


