co-organized by @christinebeck.bsky.social and Mark Adams @jacksonlab.bsky.social Register www.jax.org/longread
#RNA #genomics #bioinformatics #cancer #medicine #evolution #neuroscience

co-organized by @christinebeck.bsky.social and Mark Adams @jacksonlab.bsky.social Register www.jax.org/longread
#RNA #genomics #bioinformatics #cancer #medicine #evolution #neuroscience


Plasmid genomes were resolved using #PacBio HiFi sequencing with hifiasm-meta for #metagenome assembly. Host association was detected using epigenetic signals.
doi.org/10.1101/2025...

Plasmid genomes were resolved using #PacBio HiFi sequencing with hifiasm-meta for #metagenome assembly. Host association was detected using epigenetic signals.
doi.org/10.1101/2025...
Aardvark: Sifting through differences in a mound of variants
GitHub: github.com/PacificBiosc...
Some highlights in this thread:
1/N
Aardvark: Sifting through differences in a mound of variants
GitHub: github.com/PacificBiosc...
Some highlights in this thread:
1/N




www.nature.com/articles/s41...

www.nature.com/articles/s41...


www.hitseq.org


What’s new?
🔹 Enhanced symbolic variant support for
🔹 Robust BND comparison for cross-representation SV matching
🔹 Improved SV sequence similarity & HUGE SV support
🔹 Cleaner UI & Revamped API
👉 More: github.com/ACEnglish/tr...
#Genomics #Bioinformatics

What’s new?
🔹 Enhanced symbolic variant support for
🔹 Robust BND comparison for cross-representation SV matching
🔹 Improved SV sequence similarity & HUGE SV support
🔹 Cleaner UI & Revamped API
👉 More: github.com/ACEnglish/tr...
#Genomics #Bioinformatics
doi.org/10.1093/bioi...

doi.org/10.1093/bioi...
www.biorxiv.org/content/10.1...
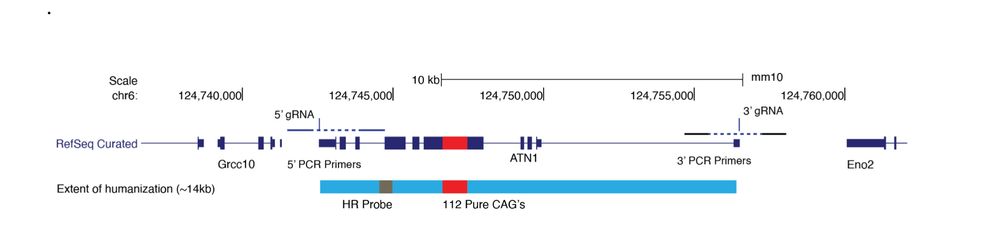
www.biorxiv.org/content/10.1...

www.nature.com/articles/s41...

www.nature.com/articles/s41...

www.nature.com/articles/s41...
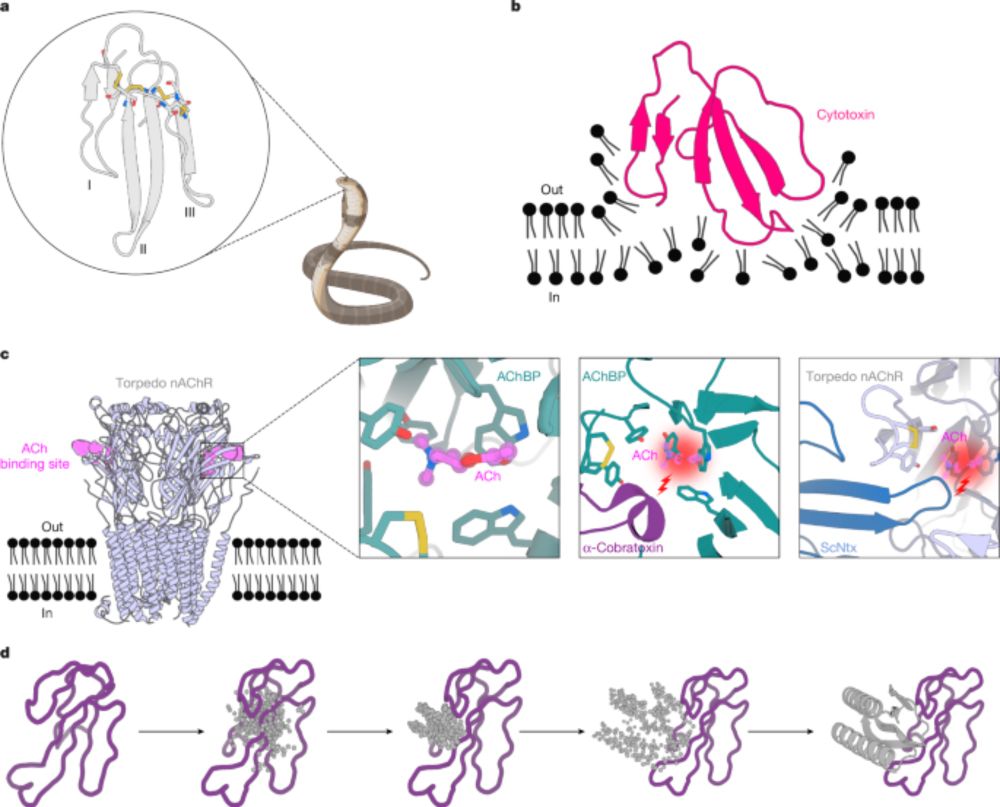
www.nature.com/articles/s41...


