
📄 Paper: www.nature.com/articles/s41...
💻 Weights: github.com/google-deepm...
Getting here wasn’t a straight path. We discussed the story behind the model, paper & API in the following roundtable: youtu.be/V8lhUqKqzUc

📄 Paper: www.nature.com/articles/s41...
💻 Weights: github.com/google-deepm...
Getting here wasn’t a straight path. We discussed the story behind the model, paper & API in the following roundtable: youtu.be/V8lhUqKqzUc

First authors: @saracarioscia.bsky.social & @aabiddanda.github.io

First authors: @saracarioscia.bsky.social & @aabiddanda.github.io
employment.utah.edu/salt-lake-ci...
employment.utah.edu/salt-lake-ci...

1/n




Grateful to all those who've been a part of this journey so far. But the fun is just getting started 💪
Curious to find out just what this privilege of tenure can enable. Let's see how we can put it to the test. Feel free to share ideas 😉
#AcademicSky

Grateful to all those who've been a part of this journey so far. But the fun is just getting started 💪
Curious to find out just what this privilege of tenure can enable. Let's see how we can put it to the test. Feel free to share ideas 😉
#AcademicSky
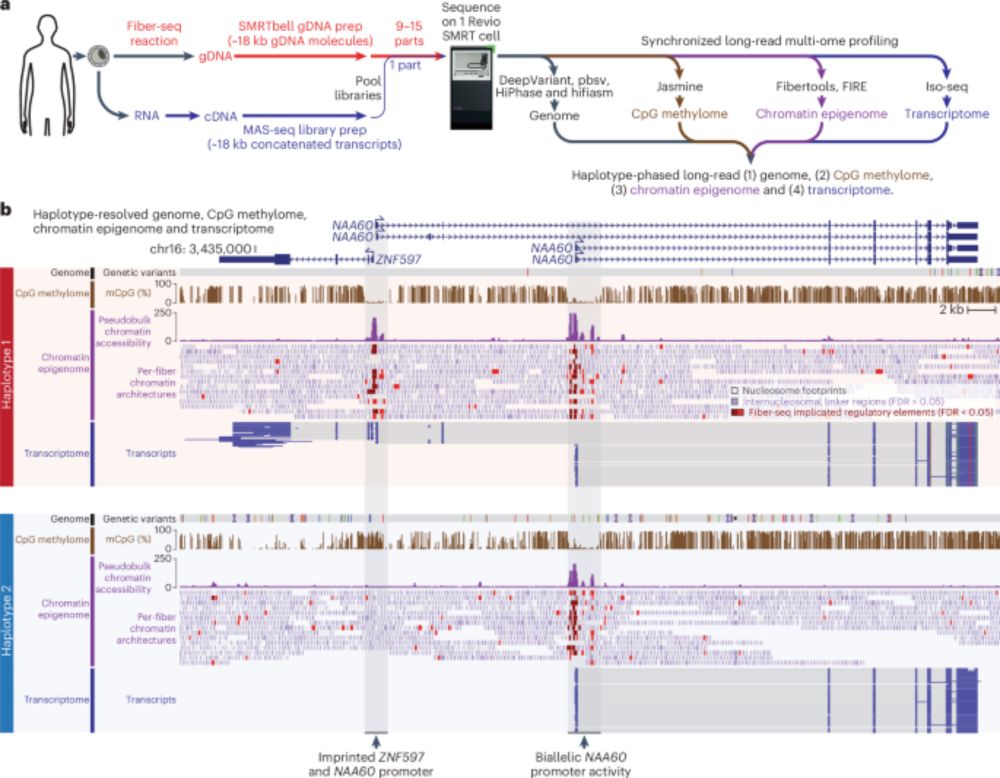
It's a very cool method for adding chromatin accessibility data to native LRS experiments.
You should definitely check out the original paper: www.science.org/doi/10.1126/...

It's a very cool method for adding chromatin accessibility data to native LRS experiments.
You should definitely check out the original paper: www.science.org/doi/10.1126/...
apnews.com/article/nih-...
apnews.com/article/nih-...
Again top-notch science with great presentations and style of bring complex matters to a broad audience! What a treat!
Thanks @mrvollger.bsky.social; @elfridedebaere.bsky.social and Musa Mhlanga @eshg.bsky.social
Again top-notch science with great presentations and style of bring complex matters to a broad audience! What a treat!
Thanks @mrvollger.bsky.social; @elfridedebaere.bsky.social and Musa Mhlanga @eshg.bsky.social

www.nature.com/articles/s41...
www.nature.com/articles/s41...

www.nature.com/articles/s41...

