
- paper: openreview.net/forum?id=WTI... !
- benchmarking platform: openproblems.bio/results/pert...
🧵1/8
Grateful to Cellarity and @fabiantheis.bsky.social for the opportunity to contribute to this outstanding project!

Grateful to Cellarity and @fabiantheis.bsky.social for the opportunity to contribute to this outstanding project!


It's my first time on the West Coast - If you are around and would like to talk about ML and/or biology, hit me up!
Looking fwd to the AI x Bio Unconference tomorrow 🚀

It's my first time on the West Coast - If you are around and would like to talk about ML and/or biology, hit me up!
Looking fwd to the AI x Bio Unconference tomorrow 🚀
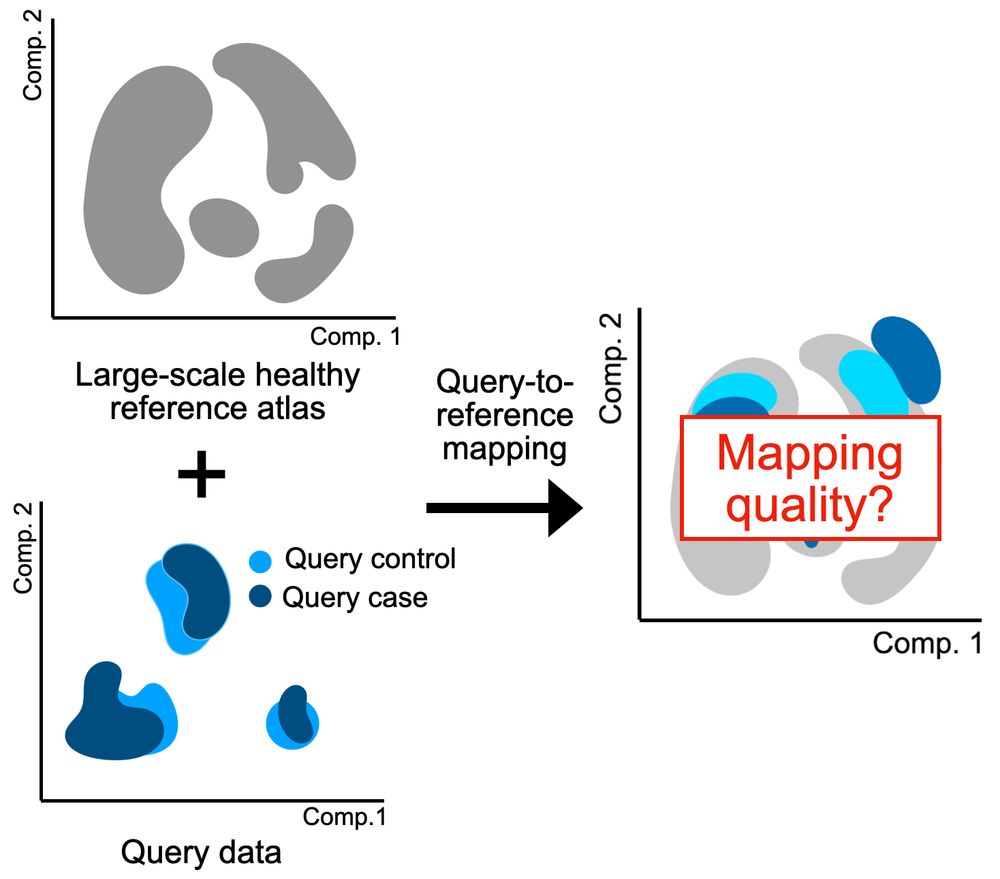
Built on flow matching, CellFlow can help guide your next phenotypic screen: biorxiv.org/content/10.1101/2025.04.11.648220v1
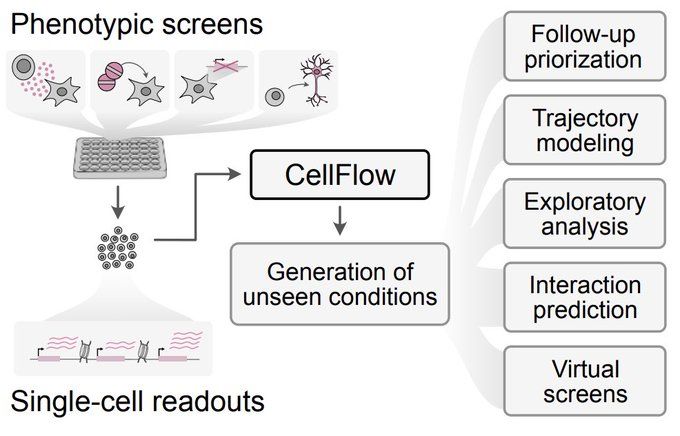
Built on flow matching, CellFlow can help guide your next phenotypic screen: biorxiv.org/content/10.1101/2025.04.11.648220v1
Keep reading for more about how we did the study and what we found out 🧵 👇
1/16
Keep reading for more about how we did the study and what we found out 🧵 👇
1/16
If you're looking for a complimentary scRNA-seq drug perturbations in healthy/primary tissue (PBMCs), check out our dataset with ~36% # drugs of Tahoe. proceedings.neurips.cc/paper_files/...
If you're looking for a complimentary scRNA-seq drug perturbations in healthy/primary tissue (PBMCs), check out our dataset with ~36% # drugs of Tahoe. proceedings.neurips.cc/paper_files/...
Hugging Face is openly reproducing the pipeline of 🐳 DeepSeek-R1. Open data, open training. open models, open collaboration.
🫵 Let's go!
github.com/huggingface/...
Hugging Face is openly reproducing the pipeline of 🐳 DeepSeek-R1. Open data, open training. open models, open collaboration.
🫵 Let's go!
github.com/huggingface/...


biorxiv.org/content/10.1101/2024.12.11.627935v1
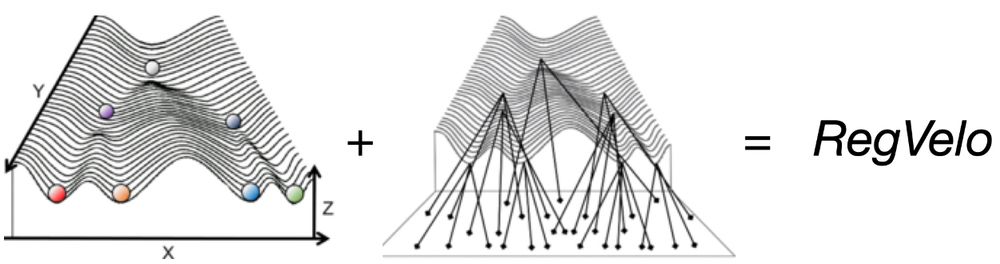
biorxiv.org/content/10.1101/2024.12.11.627935v1
@chatgtp.bsky.social
neurips.cc/virtual/2024...
@chatgtp.bsky.social
neurips.cc/virtual/2024...

