

Here we reveal an exceptional diversity of viral 2H phosphodiesterases (PDEs) that enable immune evasion by selectively degrading oligonucleotide-based messengers. This 2H PDE fold has evolved striking substrate breath & specificity.

Here we reveal an exceptional diversity of viral 2H phosphodiesterases (PDEs) that enable immune evasion by selectively degrading oligonucleotide-based messengers. This 2H PDE fold has evolved striking substrate breath & specificity.
www.nature.com/articles/s41...

www.nature.com/articles/s41...
🔗 www.pnas.org/doi/10.1073/...
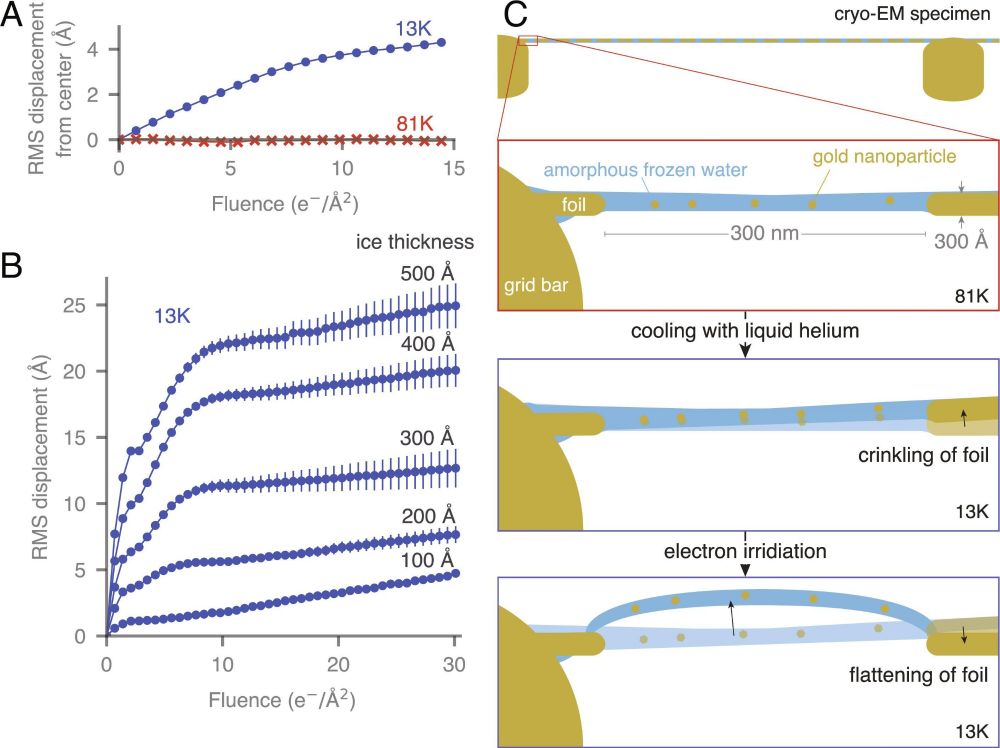
🔗 www.pnas.org/doi/10.1073/...
📄 www.biorxiv.org/content/10.1...
💾 github.com/steineggerla...
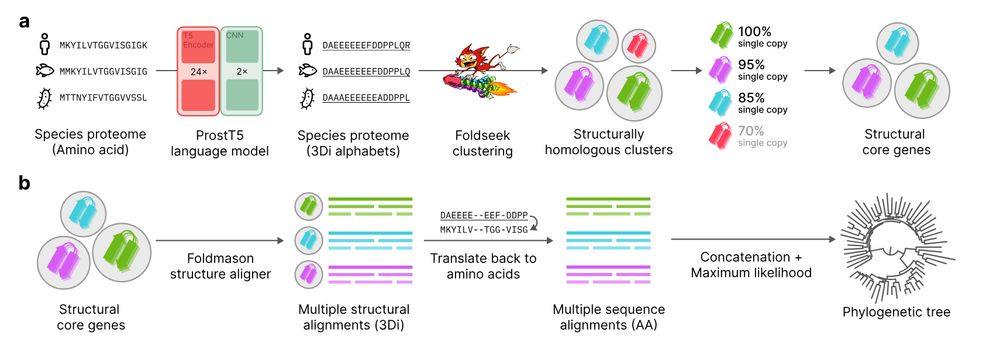
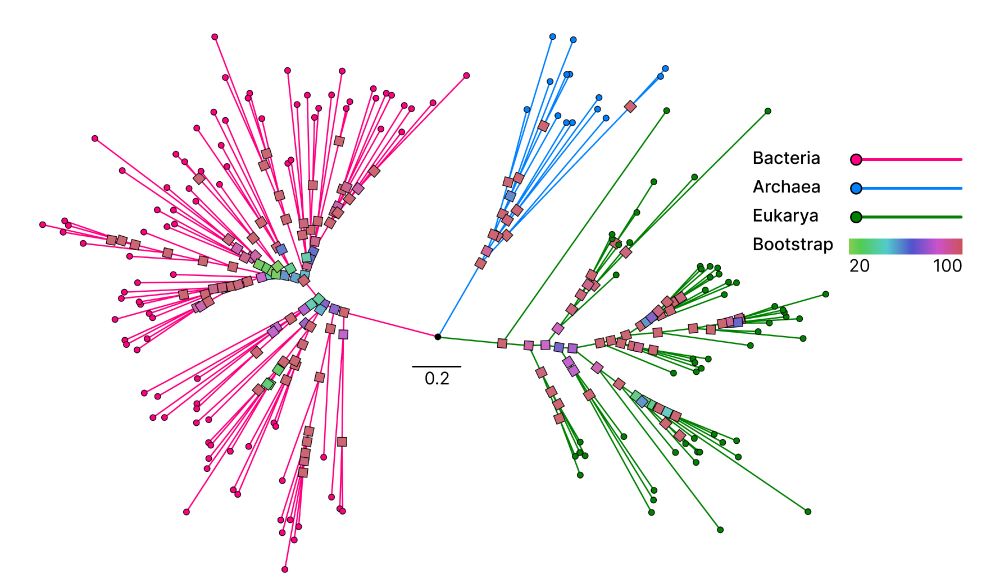
📄 www.biorxiv.org/content/10.1...
💾 github.com/steineggerla...
AI is persuasive – and able to spit out a lot of content quickly – so going to it first can severely constrain your ability to think differently. @emollick.bsky.social also talked about this on the @nightsciencepod.bsky.social.
AI is persuasive – and able to spit out a lot of content quickly – so going to it first can severely constrain your ability to think differently. @emollick.bsky.social also talked about this on the @nightsciencepod.bsky.social.
@cellpress.bsky.social
@xiaoyan52802927.bsky.social
www.cell.com/cell/fulltex...
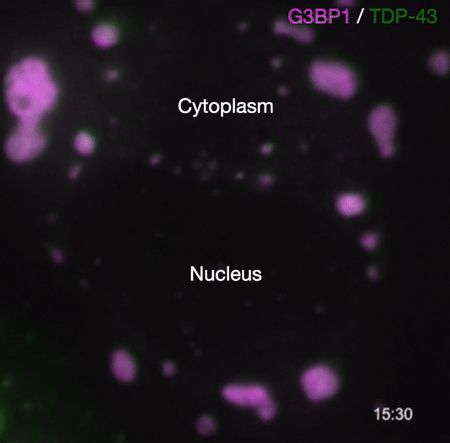
Demixing of TDP-43 inside stress granules generates pathological aggregates linked to ALS/FTD
@mpi-cbg.de @biotec-tud.bsky.social
@cellpress.bsky.social
@xiaoyan52802927.bsky.social
www.cell.com/cell/fulltex...
Demixing of TDP-43 inside stress granules generates pathological aggregates linked to ALS/FTD
@mpi-cbg.de @biotec-tud.bsky.social
We are actively working improving/updating various aspects of FINCHES; don't hesitate to reach out if you run into issues, have questions.
www.science.org/doi/10.1126/...
We are actively working improving/updating various aspects of FINCHES; don't hesitate to reach out if you run into issues, have questions.
www.science.org/doi/10.1126/...
If structure ends up big & central, u’ll render it with PyMol et al, but for a truly schematic depiction u want a biorender-style icon, but for YOUR protein.
Think I’ve finally found a way.
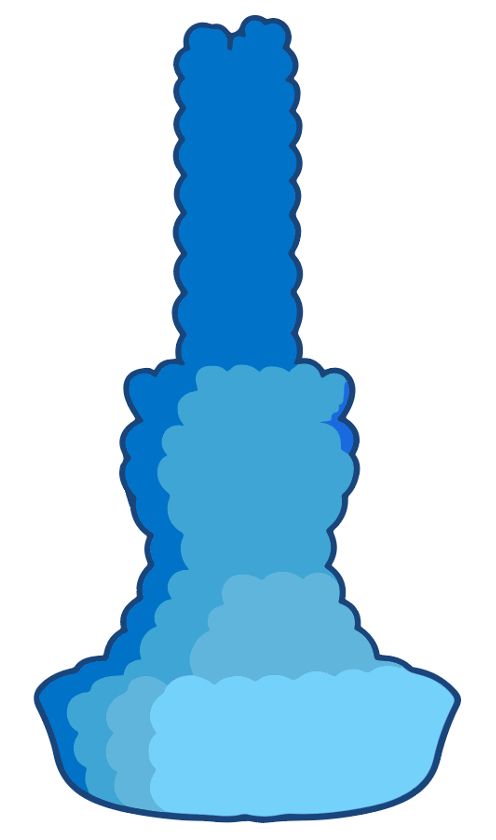
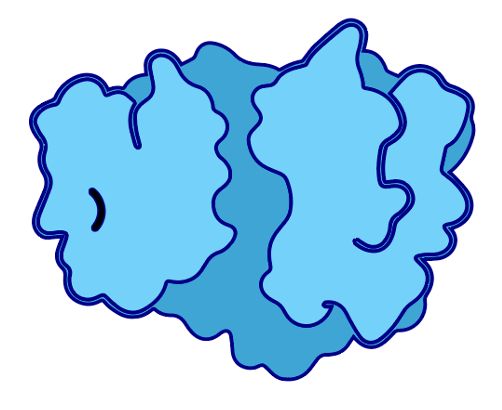

If structure ends up big & central, u’ll render it with PyMol et al, but for a truly schematic depiction u want a biorender-style icon, but for YOUR protein.
Think I’ve finally found a way.


