
So I made this video explaining how RNA velocity in situ infers gene expression dynamics by distinguishing nuclear vs. cytoplasmic expression in spatial transcriptomics data: youtu.be/z9Oxf1hvum0
#AcademicSky

So I made this video explaining how RNA velocity in situ infers gene expression dynamics by distinguishing nuclear vs. cytoplasmic expression in spatial transcriptomics data: youtu.be/z9Oxf1hvum0
#AcademicSky
---
#proteomics #prot-paper

---
#proteomics #prot-paper
---
#proteomics #prot-paper

---
#proteomics #prot-paper
Please check out all the project descriptions at the site below:
Please check out all the project descriptions at the site below:
---
#proteomics #prot-other

---
#proteomics #prot-other
---
#proteomics #prot-paper

---
#proteomics #prot-paper

We have just put this paper online: www.biorxiv.org/content/10.1...
Here, we came up with some Recommendations for Quantitative Data-Independent Acquisition (DIA) Proteomics using Controlled Quantitative Experiments (CQEs)
1/4

We have just put this paper online: www.biorxiv.org/content/10.1...
Here, we came up with some Recommendations for Quantitative Data-Independent Acquisition (DIA) Proteomics using Controlled Quantitative Experiments (CQEs)
1/4
The regulation of most proteins is dominated by different regulatory mechanisms across cell types.
Gratifyingly, this complex regulation defines simple rules ⬇️
www.biorxiv.org/content/10.1...

The regulation of most proteins is dominated by different regulatory mechanisms across cell types.
Gratifyingly, this complex regulation defines simple rules ⬇️
www.biorxiv.org/content/10.1...
---
#proteomics #prot-preprint

---
#proteomics #prot-preprint
pubs.acs.org/doi/10.1021/...
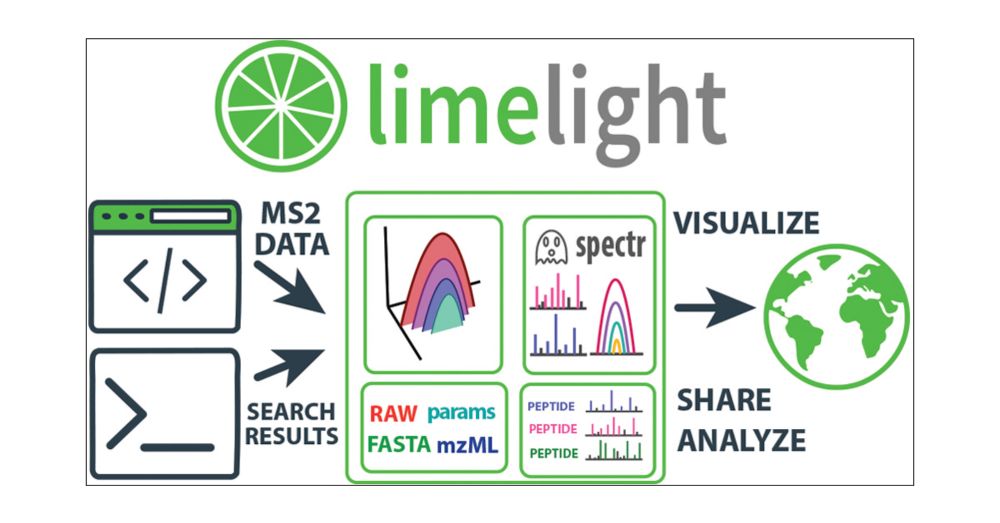
pubs.acs.org/doi/10.1021/...

---
#proteomics #prot-paper

---
#proteomics #prot-paper
---
#proteomics #prot-other
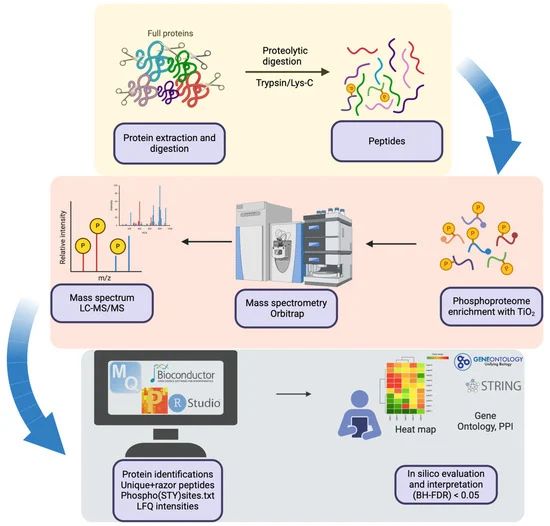
---
#proteomics #prot-other
---
#proteomics #prot-other

---
#proteomics #prot-other
Check it out:
arxiv.org/abs/2502.15867

Check it out:
arxiv.org/abs/2502.15867
#proteomics #metabolomics #lipidomics
discord.gg/Sm6gWgpsf4

#proteomics #metabolomics #lipidomics
discord.gg/Sm6gWgpsf4

#proteomics #metabolomics #lipidomics
discord.gg/Sm6gWgpsf4

The registration is open 👇nmr2025.conferences-pasteur.org

The registration is open 👇nmr2025.conferences-pasteur.org
Renew your ANZSMS membership for 2025-2026 today to take advantage of discounted registration rates for AusOMICs. www.anzsms.org/membership/
#TeamMassSpec #massspectrometry #massspec

Renew your ANZSMS membership for 2025-2026 today to take advantage of discounted registration rates for AusOMICs. www.anzsms.org/membership/
#TeamMassSpec #massspectrometry #massspec
-> go.nature.com/4gCYtuL
A huge thank to the @fendtlab.bsky.social and all our collaborators for the terrific team work 🙏
A brief 🧵
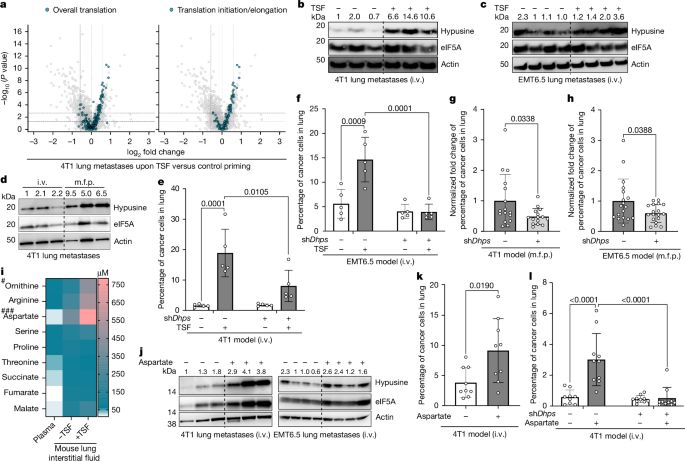
-> go.nature.com/4gCYtuL
A huge thank to the @fendtlab.bsky.social and all our collaborators for the terrific team work 🙏
A brief 🧵
---
#proteomics #prot-paper

---
#proteomics #prot-paper
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

