
biotech BSc USAL & bioinfo MSc @uv.es
💻 github.com/ahmig | ahmig.es
📍 València, Spain
now you can swap in different stats (useful for non-linear allele freq trends). also: new Docker images on release, containerized CI, updated envs, and minor bug fixes for smoother, reproducible intra-patient SARS-CoV-2 analyses
🧪👉 github.com/PathoGenOmics-Lab/VIPERA

now you can swap in different stats (useful for non-linear allele freq trends). also: new Docker images on release, containerized CI, updated envs, and minor bug fixes for smoother, reproducible intra-patient SARS-CoV-2 analyses
🧪👉 github.com/PathoGenOmics-Lab/VIPERA
This story unfolded pretty fast yesterday for a few reasons. The object was discovered on July 1st UTC by the Asteroid Terrestrial-impact Last Alert System, or ATLAS team. A #NASA funded survey at the University of Hawaii. atlas.fallingstar.com 🔭🧪
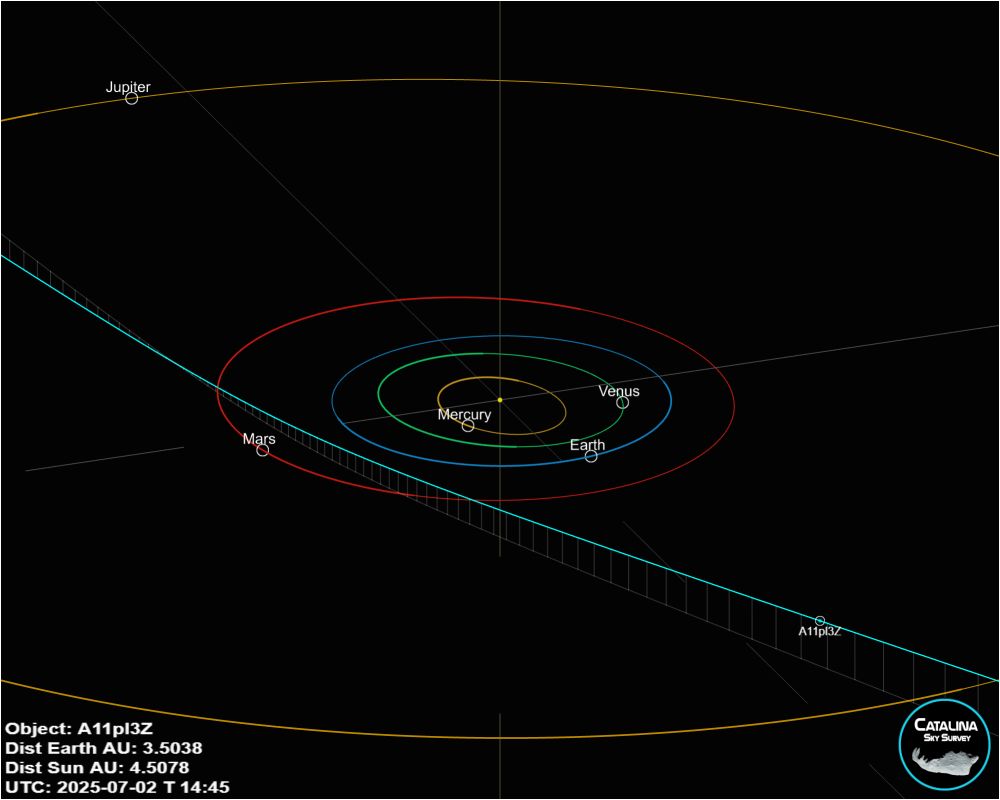
This story unfolded pretty fast yesterday for a few reasons. The object was discovered on July 1st UTC by the Asteroid Terrestrial-impact Last Alert System, or ATLAS team. A #NASA funded survey at the University of Hawaii. atlas.fallingstar.com 🔭🧪
(tagging🔭☄️)
Observing it with remote scope rn, calling astronomers..
(tagging🔭☄️)
Observing it with remote scope rn, calling astronomers..
🌐 i2sysbio.es/communicatio...

🌐 i2sysbio.es/communicatio...
Our workflow for SARS-CoV-2 intra-patient evolution analysis now fully supports Snakemake 8/9 (with dedicated CI tests), BIONJ trees, richer reports, added config parameters, modular Snakefiles and more! 🧪👉 github.com/PathoGenOmics-Lab/VIPERA

Our workflow for SARS-CoV-2 intra-patient evolution analysis now fully supports Snakemake 8/9 (with dedicated CI tests), BIONJ trees, richer reports, added config parameters, modular Snakefiles and more! 🧪👉 github.com/PathoGenOmics-Lab/VIPERA



delegacion.comunitatvalenciana.csic.es/hallan-error...

delegacion.comunitatvalenciana.csic.es/hallan-error...

while exploring how often #sars-cov-2 "repairs" deletions in BA.1, we found significant genome artifacts in GISAID 🤯 we compared with raw reads from ENA + tested the effects in vitro. turns out, data processing matters a lot 📉
check it out!! doi.org/10.1093/ve/v...

while exploring how often #sars-cov-2 "repairs" deletions in BA.1, we found significant genome artifacts in GISAID 🤯 we compared with raw reads from ENA + tested the effects in vitro. turns out, data processing matters a lot 📉
check it out!! doi.org/10.1093/ve/v...


https://go.nature.com/42tH8Ai

https://go.nature.com/42tH8Ai


