
Xinjun Zhang
@zhanglabpopgen.bsky.social
Assistant Professor at the University of Michigan. Popgen/Evolgen, admixture everything, ML etc. More on www.zhanglabpopgen.org
Reposted by Xinjun Zhang
I am thrilled to share this paper outlining some ideas I’ve been thinking about for a little while on a simple but powerful approach for predicting risk of inbreeding depression from long runs of homozygosity and non-ROH heterozygosity. 1/n @klohmueller.bsky.social doi.org/10.1016/j.tr...
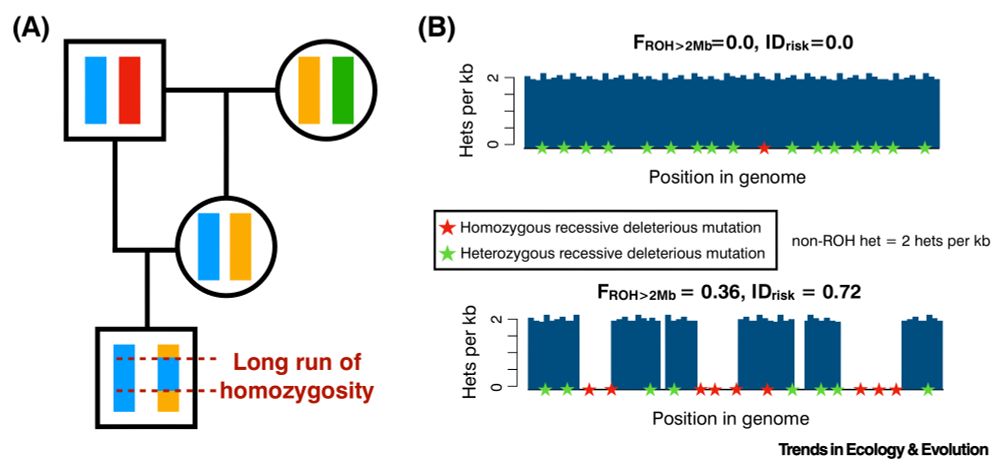
August 4, 2025 at 5:10 PM
I am thrilled to share this paper outlining some ideas I’ve been thinking about for a little while on a simple but powerful approach for predicting risk of inbreeding depression from long runs of homozygosity and non-ROH heterozygosity. 1/n @klohmueller.bsky.social doi.org/10.1016/j.tr...
Reposted by Xinjun Zhang
I am seeking a postdoc for my group at UCLA. We work at the intersection of population genetics x microbiome (garud.eeb.ucla.edu). If interested, please message me!
Garud Lab
garud.eeb.ucla.edu
July 22, 2025 at 5:51 PM
I am seeking a postdoc for my group at UCLA. We work at the intersection of population genetics x microbiome (garud.eeb.ucla.edu). If interested, please message me!
Reposted by Xinjun Zhang
Sophie Buysse's paper on short stamen loss in Arabipsis is published in this month's Evolution issue!! academic.oup.com/evolut/artic... @journal-evo.bsky.social
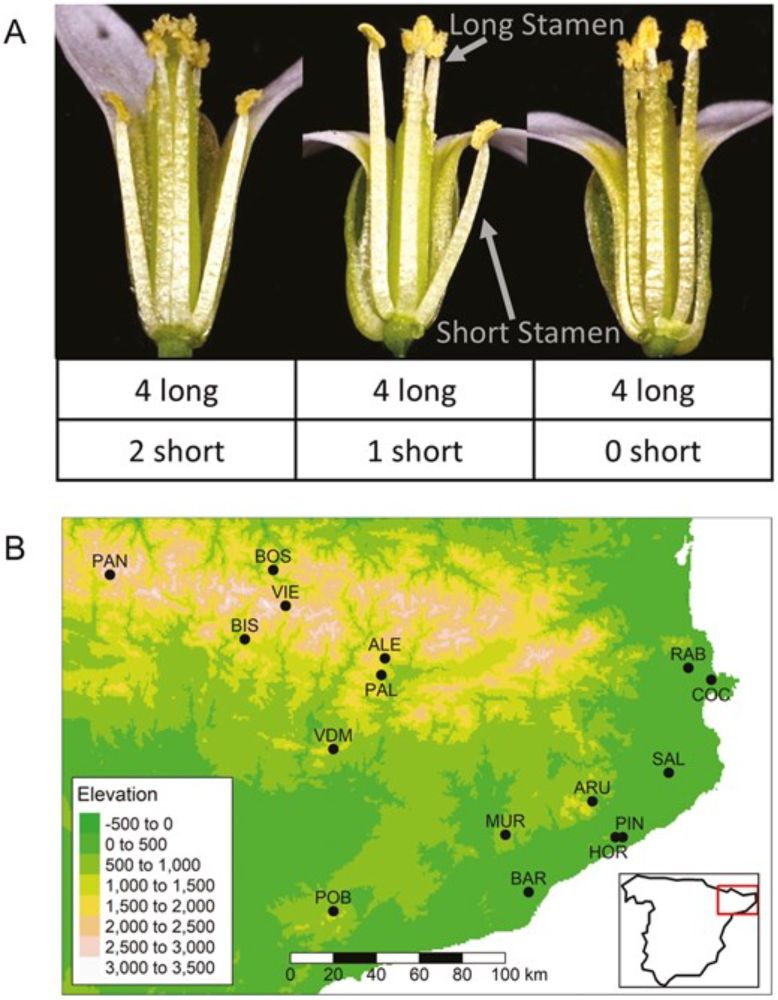
Evaluating the roles of drift and selection in trait loss along an elevational gradient
The evolutionary mechanisms underlying loss or retention of traits that have lost function are poorly understood. Short stamens in Arabidopsis thaliana pro
academic.oup.com
July 21, 2025 at 5:51 PM
Sophie Buysse's paper on short stamen loss in Arabipsis is published in this month's Evolution issue!! academic.oup.com/evolut/artic... @journal-evo.bsky.social
Having major FOMO about #SMBE2025 in my hometown… but I’m very proud that my postdoc, Susanna Gutierrez, will be presenting our work on inferring the selection history of adaptive introgression candidates in Peruvians at poster S12-P19. Please visit her during poster session 3 if you're interested!
July 21, 2025 at 6:13 AM
Having major FOMO about #SMBE2025 in my hometown… but I’m very proud that my postdoc, Susanna Gutierrez, will be presenting our work on inferring the selection history of adaptive introgression candidates in Peruvians at poster S12-P19. Please visit her during poster session 3 if you're interested!
Reposted by Xinjun Zhang
Reposted by Xinjun Zhang
The preliminary program of ProbGen 2026, which will be held at UC Berkeley, is now up: probgen2026.github.io
Sharing on behalf of Rasmus Nielsen who is not on this site. For more see his thread on that other site: x.com/ras_nielsen/...
Sharing on behalf of Rasmus Nielsen who is not on this site. For more see his thread on that other site: x.com/ras_nielsen/...
Home - ProbGen 2026
Your Site Description
probgen2026.github.io
June 5, 2025 at 8:29 PM
The preliminary program of ProbGen 2026, which will be held at UC Berkeley, is now up: probgen2026.github.io
Sharing on behalf of Rasmus Nielsen who is not on this site. For more see his thread on that other site: x.com/ras_nielsen/...
Sharing on behalf of Rasmus Nielsen who is not on this site. For more see his thread on that other site: x.com/ras_nielsen/...
Super excited to share my latest preprint with @klohmueller.bsky.social! Here we leverage the unique signature that recessive deleterious variants can lead to an increase in archaic ancestry to tackle a classic question - the dominance distribution on the human genome www.biorxiv.org/content/10.1...

Neanderthal introgressed ancestry reveals human genomic regions enriched with recessive deleterious mutations
Negative natural selection on deleterious mutations plays a key role in shaping human genetic variation. Understanding the dominance of deleterious mutations is critical as it can fundamentally impact...
www.biorxiv.org
May 8, 2025 at 2:58 PM
Super excited to share my latest preprint with @klohmueller.bsky.social! Here we leverage the unique signature that recessive deleterious variants can lead to an increase in archaic ancestry to tackle a classic question - the dominance distribution on the human genome www.biorxiv.org/content/10.1...
Reposted by Xinjun Zhang
Repost w/ correct link!
The Dept of Human Genetics at the Univeristy of Utah is hosting a Rising Stars in Genetics & Genomics postdoc symposium. We are looking to feature postdocs doing cool science! Please self nominate or nominate an excellent postdoc! #ASHGtrainees docs.google.com/forms/d/e/1F...
The Dept of Human Genetics at the Univeristy of Utah is hosting a Rising Stars in Genetics & Genomics postdoc symposium. We are looking to feature postdocs doing cool science! Please self nominate or nominate an excellent postdoc! #ASHGtrainees docs.google.com/forms/d/e/1F...

May 3, 2025 at 4:50 AM
Repost w/ correct link!
The Dept of Human Genetics at the Univeristy of Utah is hosting a Rising Stars in Genetics & Genomics postdoc symposium. We are looking to feature postdocs doing cool science! Please self nominate or nominate an excellent postdoc! #ASHGtrainees docs.google.com/forms/d/e/1F...
The Dept of Human Genetics at the Univeristy of Utah is hosting a Rising Stars in Genetics & Genomics postdoc symposium. We are looking to feature postdocs doing cool science! Please self nominate or nominate an excellent postdoc! #ASHGtrainees docs.google.com/forms/d/e/1F...
Reposted by Xinjun Zhang
Kirk Lohmueller
DominL uses patterns of archaic ancestry to infer degree of dominance. Provides statistical support for the presence of recessive deleterious mutations in the human genome. Up to 15% of the human genome may carry recessive deleterious mutations! 🧬
#HumanEvo25
DominL uses patterns of archaic ancestry to infer degree of dominance. Provides statistical support for the presence of recessive deleterious mutations in the human genome. Up to 15% of the human genome may carry recessive deleterious mutations! 🧬
#HumanEvo25

April 29, 2025 at 3:08 PM
Kirk Lohmueller
DominL uses patterns of archaic ancestry to infer degree of dominance. Provides statistical support for the presence of recessive deleterious mutations in the human genome. Up to 15% of the human genome may carry recessive deleterious mutations! 🧬
#HumanEvo25
DominL uses patterns of archaic ancestry to infer degree of dominance. Provides statistical support for the presence of recessive deleterious mutations in the human genome. Up to 15% of the human genome may carry recessive deleterious mutations! 🧬
#HumanEvo25
Reposted by Xinjun Zhang
a quick note on my paper with @jeffspence.bsky.social and @jkpritch.bsky.social on conditional frequency spectra, now out in a @genetics-gsa.bsky.social special issue: doi.org/10.1093/genetics/iyae210
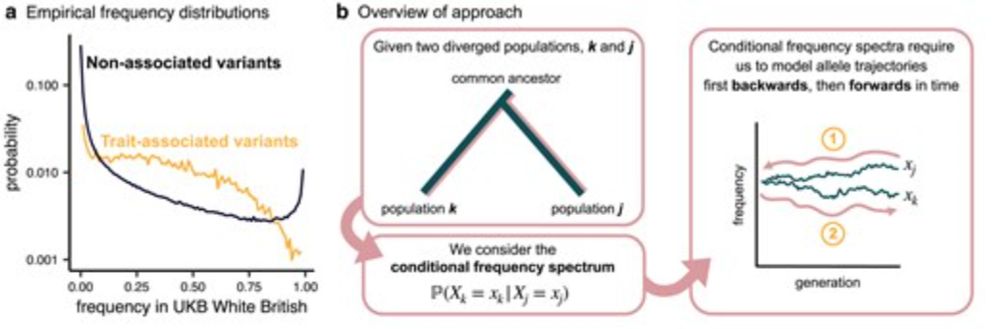
Characterizing selection on complex traits through conditional frequency spectra
Abstract. Natural selection on complex traits is difficult to study in part due to the ascertainment inherent to genome-wide association studies (GWAS). Th
doi.org
April 18, 2025 at 7:49 PM
a quick note on my paper with @jeffspence.bsky.social and @jkpritch.bsky.social on conditional frequency spectra, now out in a @genetics-gsa.bsky.social special issue: doi.org/10.1093/genetics/iyae210
Reposted by Xinjun Zhang
Excited to share: "Addressing missing context in regulatory variation across primate evolution"
arxiv.org/abs/2504.02081
arxiv.org/abs/2504.02081
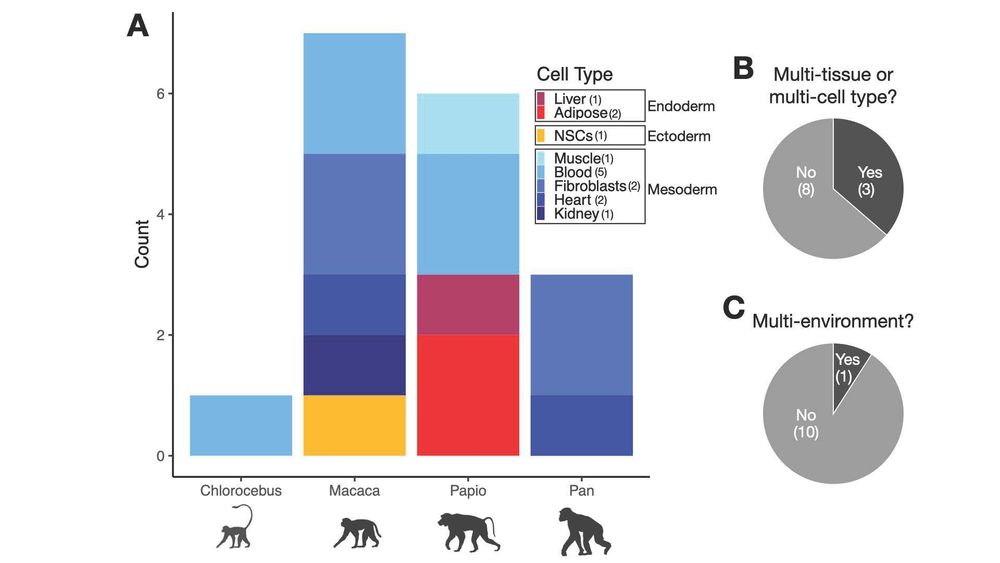
April 4, 2025 at 2:52 PM
Excited to share: "Addressing missing context in regulatory variation across primate evolution"
arxiv.org/abs/2504.02081
arxiv.org/abs/2504.02081
Reposted by Xinjun Zhang
A really nice paper by @drghawkes.bsky.social et al. argues that rare and common genetic associations converge on the same genes.
While this seems at odds with our recent work about how burden tests and GWAS prioritize different genes, our results agree (🧬🧪🧵 1/6)
www.biorxiv.org/content/10.1...
While this seems at odds with our recent work about how burden tests and GWAS prioritize different genes, our results agree (🧬🧪🧵 1/6)
www.biorxiv.org/content/10.1...

Whole-genome sequencing analysis of anthropometric traits in 672,976 individuals reveals convergence between rare and common genetic associations
Genetic association studies have mostly focussed on common variants from genotyping arrays or rare protein-coding variants from exome sequencing. Here, we used whole-genome sequence (WGS) data in 672,...
www.biorxiv.org
March 28, 2025 at 1:22 AM
A really nice paper by @drghawkes.bsky.social et al. argues that rare and common genetic associations converge on the same genes.
While this seems at odds with our recent work about how burden tests and GWAS prioritize different genes, our results agree (🧬🧪🧵 1/6)
www.biorxiv.org/content/10.1...
While this seems at odds with our recent work about how burden tests and GWAS prioritize different genes, our results agree (🧬🧪🧵 1/6)
www.biorxiv.org/content/10.1...
Reposted by Xinjun Zhang
Really excited to see this work, led by Mike Grundler, published! It's a new method for inferring the geographic locations of shared genetic ancestors. In the paper, we and use it to infer the geographic history of human genetic ancestry
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
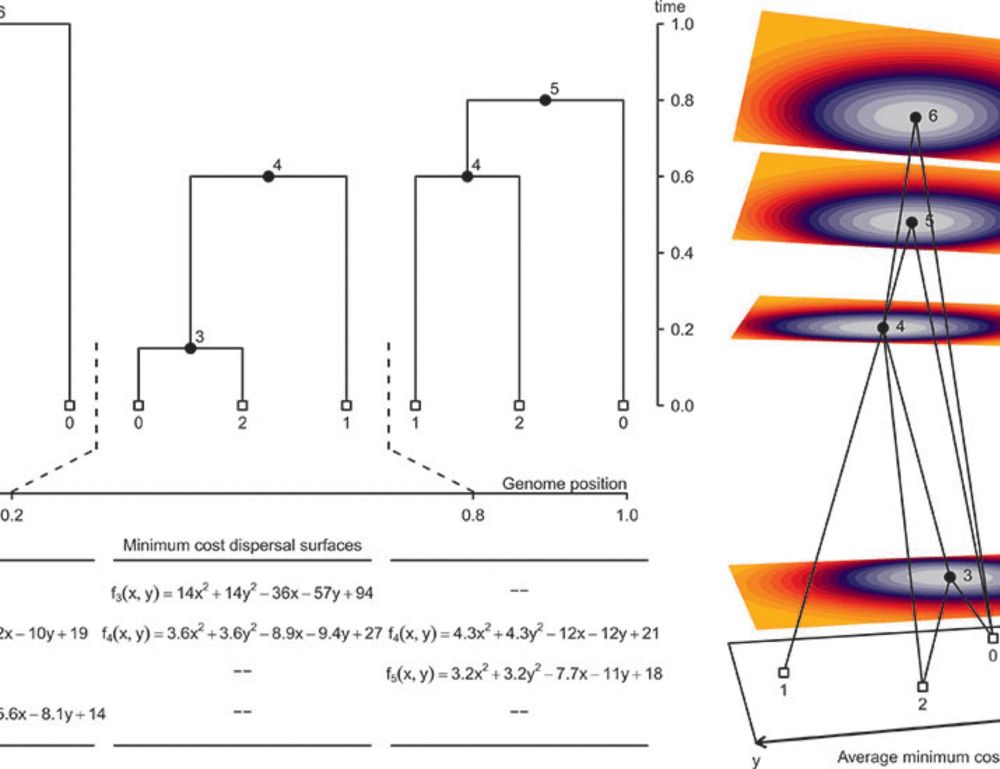
A geographic history of human genetic ancestry
Describing the distribution of genetic variation across individuals is a fundamental goal of population genetics. We present a method that capitalizes on the rich genealogical information encoded in g...
www.science.org
March 28, 2025 at 5:26 PM
Really excited to see this work, led by Mike Grundler, published! It's a new method for inferring the geographic locations of shared genetic ancestors. In the paper, we and use it to infer the geographic history of human genetic ancestry
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
Reposted by Xinjun Zhang
Grundler et al propose a new method to infer the geographic position of all our genetic ancestors www.science.org/doi/10.1126/...
building on a method by Wohns et al www.science.org/doi/10.1126/....
My thoughts on the approach: www.science.org/doi/epdf/10.....
building on a method by Wohns et al www.science.org/doi/10.1126/....
My thoughts on the approach: www.science.org/doi/epdf/10.....
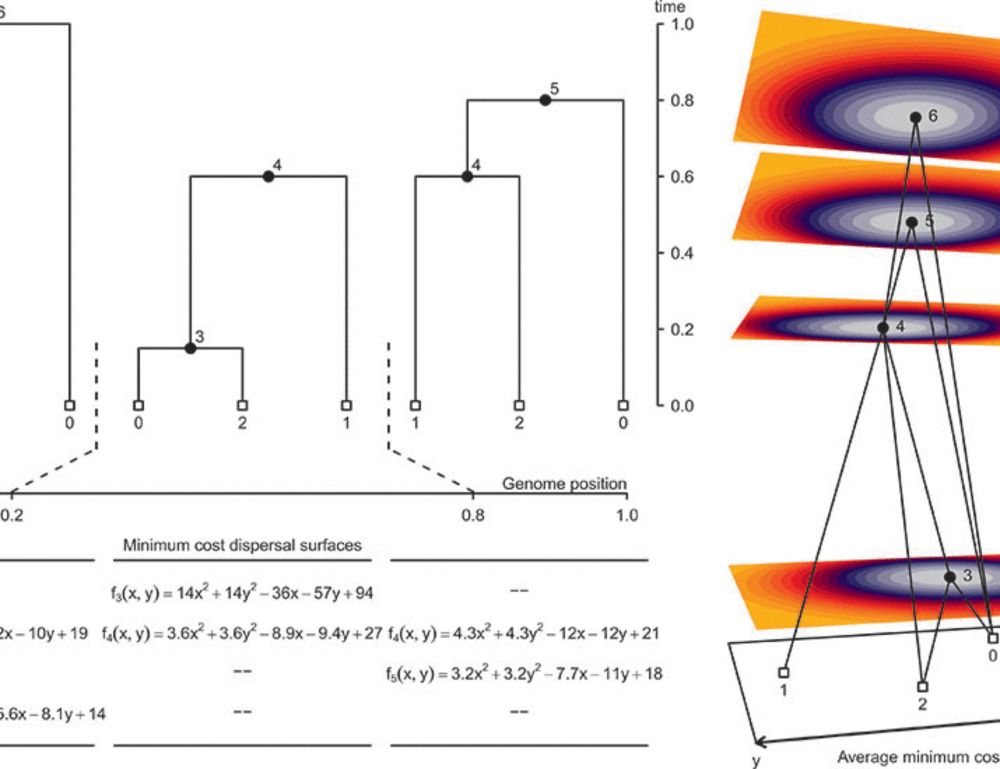
A geographic history of human genetic ancestry
Describing the distribution of genetic variation across individuals is a fundamental goal of population genetics. We present a method that capitalizes on the rich genealogical information encoded in g...
www.science.org
March 28, 2025 at 2:53 PM
Grundler et al propose a new method to infer the geographic position of all our genetic ancestors www.science.org/doi/10.1126/...
building on a method by Wohns et al www.science.org/doi/10.1126/....
My thoughts on the approach: www.science.org/doi/epdf/10.....
building on a method by Wohns et al www.science.org/doi/10.1126/....
My thoughts on the approach: www.science.org/doi/epdf/10.....
Reposted by Xinjun Zhang
Think of a polygenic score you care about. Are direct genetic effects driving variation among people in this predictor? Or perhaps other, confounding factors? We at the @arbelharpak.bsky.social & @docedge.bsky.social Labs developed a method to tackle this question. [1/n]
A Litmus Test for Confounding in Polygenic Scores
Polygenic scores (PGSs) are being rapidly adopted for trait prediction in the clinic and beyond. PGSs are often thought of as capturing the direct genetic effect of one's genotype on their phenotype. ...
www.biorxiv.org
February 4, 2025 at 6:04 PM
Think of a polygenic score you care about. Are direct genetic effects driving variation among people in this predictor? Or perhaps other, confounding factors? We at the @arbelharpak.bsky.social & @docedge.bsky.social Labs developed a method to tackle this question. [1/n]
Reposted by Xinjun Zhang
We are thrilled to share that our paper entitled "Inference of the demographic histories and selective effects of human gut commensal microbiota over the course of human history" is officially out in MBE! academic.oup.com/mbe/advance-...

Inference of the demographic histories and selective effects of human gut commensal microbiota over the course of human history
Abstract. Despite the importance of gut commensal microbiota to human health, there is little knowledge about their evolutionary histories, including their
academic.oup.com
January 24, 2025 at 6:14 PM
We are thrilled to share that our paper entitled "Inference of the demographic histories and selective effects of human gut commensal microbiota over the course of human history" is officially out in MBE! academic.oup.com/mbe/advance-...
Reposted by Xinjun Zhang
Thrilled to have been a part of this study led by @aidaandres.bsky.social that finds evidence of genetic adaptation in chimpanzees to different habitats. Most notably signatures of positive selection in chimpanzees underlie resistance to malaria in humans (GYPA and HBB).
Chimpanzees bear genetic adaptations that help them thrive in their forest and savannah habitats, some of which may protect against malaria: new @science.org paper led by Prof @aidaandres.bsky.social & Harrison Ostridge @ugiatucl.bsky.social @ucllifesciences.bsky.social www.ucl.ac.uk/news/2025/ja...

Chimpanzees are genetically adapted to local habitats and infections such as malaria
Chimpanzees bear genetic adaptations that help them thrive in their different forest and savannah habitats, some of which may protect against malaria, according to a study by an international team led...
www.ucl.ac.uk
January 10, 2025 at 5:29 PM
Thrilled to have been a part of this study led by @aidaandres.bsky.social that finds evidence of genetic adaptation in chimpanzees to different habitats. Most notably signatures of positive selection in chimpanzees underlie resistance to malaria in humans (GYPA and HBB).


