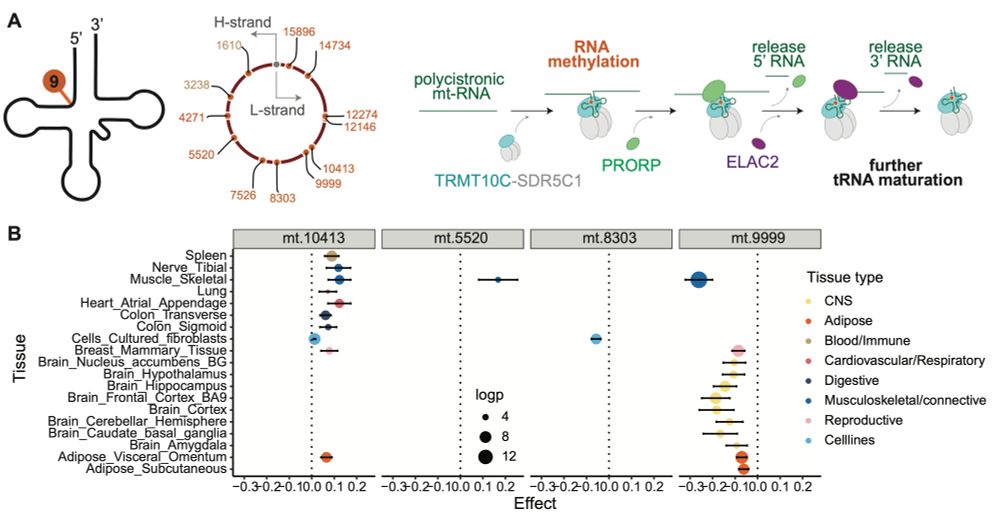
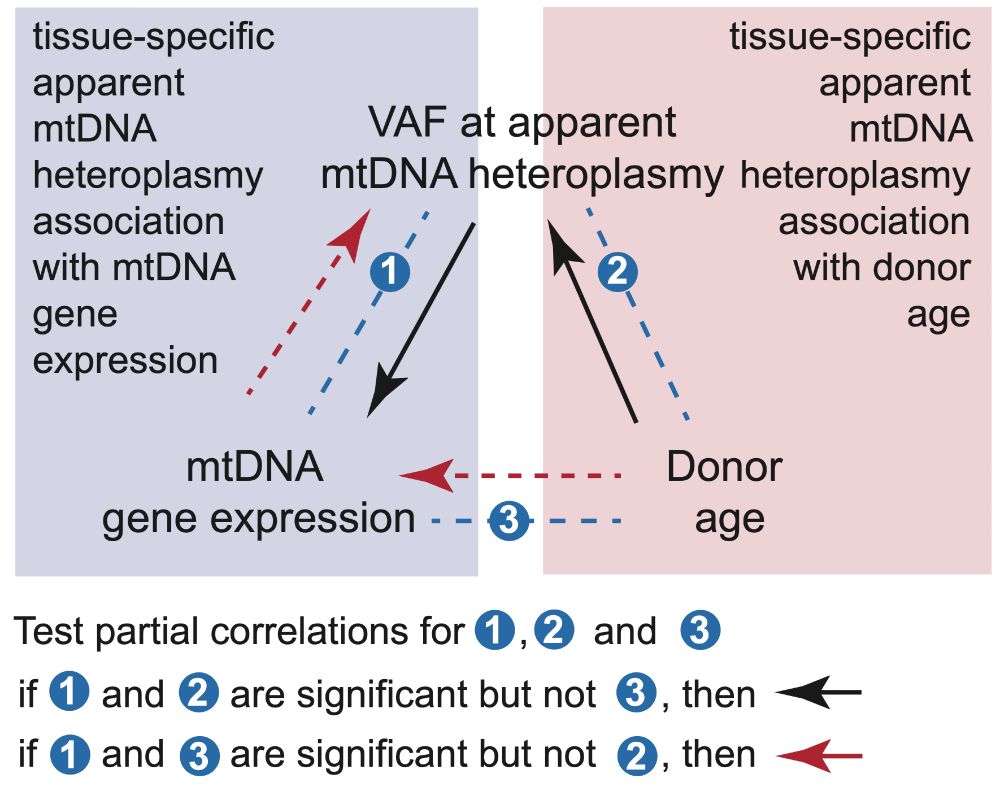
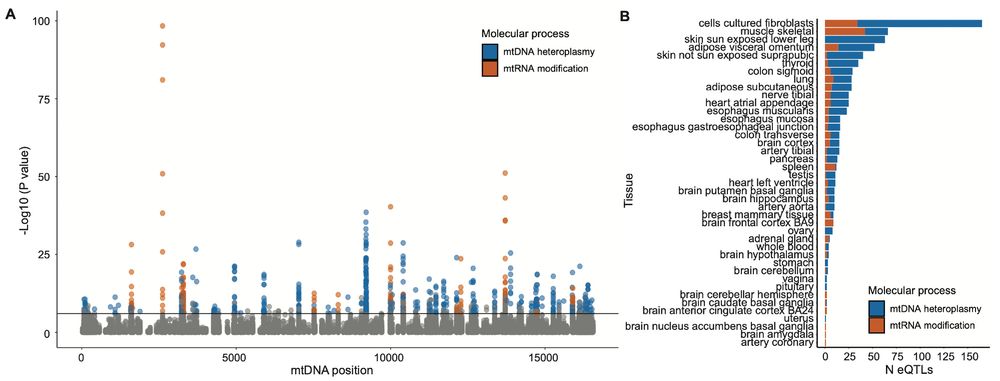
Our latest work on causal models for this is out yesterday:
www.nature.com/articles/s41...
A short🧵:

Our latest work on causal models for this is out yesterday:
www.nature.com/articles/s41...
A short🧵:
I will record lectures & all will be found at this link: github.com/rmcelreath/s...

I will record lectures & all will be found at this link: github.com/rmcelreath/s...
3 years funding, self-designed interdisciplinary project co-led between EMBL PI and PI in one of EMBL's actual, prospect or associate member states.
In particular: AI&Theory, use mathem. models, physics principles and AI
www.embl.org/about/info/p...
3 years funding, self-designed interdisciplinary project co-led between EMBL PI and PI in one of EMBL's actual, prospect or associate member states.
In particular: AI&Theory, use mathem. models, physics principles and AI
www.embl.org/about/info/p...
I am excited to announce that our new study explaining the missing heritability of many phenotypes using WGS data from ~347,000 UK Biobank participants has just been published in @Nature.
Our manuscript is here: www.nature.com/articles/s41....

I am excited to announce that our new study explaining the missing heritability of many phenotypes using WGS data from ~347,000 UK Biobank participants has just been published in @Nature.
Our manuscript is here: www.nature.com/articles/s41....
1/n

1/n
You'll be working on multi-omics factorization models for personalized medicine, co-supervised by @junyanlu.bsky.social.
👉 Project: www.helmholtz.de/assets/hidss...
✍️ Apply: hidss4health.de/application

You'll be working on multi-omics factorization models for personalized medicine, co-supervised by @junyanlu.bsky.social.
👉 Project: www.helmholtz.de/assets/hidss...
✍️ Apply: hidss4health.de/application
@medrxivpreprint.bsky.social! This huge collaborative effort advances our understanding of psychiatric genetic architecture and emphasizes the importance of looking beyond additive effects. 🧵1/n; 🧪👩🏽🔬🧬
@medrxivpreprint.bsky.social! This huge collaborative effort advances our understanding of psychiatric genetic architecture and emphasizes the importance of looking beyond additive effects. 🧵1/n; 🧪👩🏽🔬🧬

www.uu.se/en/about-uu/...

www.uu.se/en/about-uu/...
1/n
1/n
@caina89.bsky.social !!! Thank you for this amazing journey!!! 🤩
@caina89.bsky.social !!! Thank you for this amazing journey!!! 🤩