Viruses | Immunology | Ribosomes | Systems Biology
#LoveVirology
https://www.shiraweingartengabbay.com/
rdcu.be/e1bBW

rdcu.be/e1bBW
Paper: pubmed.ncbi.nlm.nih.gov/40263451/
#BillSchneider and I are delighted to announce another round of cutting-edge talks in #SystemsVirology! 🦠💡
Join us and an outstanding lineup of speakers, starting Oct 30.
Free registration: shiraweingartengabbay.com/systems-viro... 🔬✨

Paper: pubmed.ncbi.nlm.nih.gov/40263451/
#BillSchneider and I are delighted to announce another round of cutting-edge talks in #SystemsVirology! 🦠💡
Join us and an outstanding lineup of speakers, starting Oct 30.
Free registration: shiraweingartengabbay.com/systems-viro... 🔬✨

Dr. @shiraweingarten.bsky.social at @harvardmed.bsky.social launched the Laboratory of Systems Virology. We cannot wait to see what the lab accomplishes in 2026!
https://bsky.app/profile/scienceinboston.com/post/3lsuek25o5c2f
Her lab uncovers the inner workings of viruses and their interaction with the immune system to prevent current and future viral diseases: bit.ly/44peaBa

Dr. @shiraweingarten.bsky.social at @harvardmed.bsky.social launched the Laboratory of Systems Virology. We cannot wait to see what the lab accomplishes in 2026!
https://bsky.app/profile/scienceinboston.com/post/3lsuek25o5c2f
www.nature.com/articles/d41...

www.nature.com/articles/d41...
@drsprggs.bsky.social and @tylernstarr.bsky.social. Check your email for registration link and form to submit questions in advance.

@drsprggs.bsky.social and @tylernstarr.bsky.social. Check your email for registration link and form to submit questions in advance.
Paper: pubmed.ncbi.nlm.nih.gov/40345199/
#BillSchneider and I are delighted to announce another round of cutting-edge talks in #SystemsVirology! 🦠💡
Join us and an outstanding lineup of speakers, starting Oct 30.
Free registration: shiraweingartengabbay.com/systems-viro... 🔬✨

Paper: pubmed.ncbi.nlm.nih.gov/40345199/
Paper: pubmed.ncbi.nlm.nih.gov/38723628/
#BillSchneider and I are delighted to announce another round of cutting-edge talks in #SystemsVirology! 🦠💡
Join us and an outstanding lineup of speakers, starting Oct 30.
Free registration: shiraweingartengabbay.com/systems-viro... 🔬✨

Paper: pubmed.ncbi.nlm.nih.gov/38723628/
#BillSchneider and I are delighted to announce another round of cutting-edge talks in #SystemsVirology! 🦠💡
Join us and an outstanding lineup of speakers, starting Oct 30.
Free registration: shiraweingartengabbay.com/systems-viro... 🔬✨

#BillSchneider and I are delighted to announce another round of cutting-edge talks in #SystemsVirology! 🦠💡
Join us and an outstanding lineup of speakers, starting Oct 30.
Free registration: shiraweingartengabbay.com/systems-viro... 🔬✨
@sohailtavazoie.bsky.social's lab, and Maria Cecilia Campos Canesso, formerly a postdoc in @danmucida.bsky.social and @victora.bsky.social’s labs. A very well-deserved recognition!

🔸Farren Isaacs
🔸Jessica Stark @jcstark.bsky.social
🔸Shira Weingarten-Gabbay @shiraweingarten.bsky.social
🔸Abhishek Chatterjee @achemsynbio.bsky.social
🔸Wen Tseng
🔸Michael Krogh Jensen
🔸Ryan Clarke
🔸Alex Reis @alexcampreis.com

Our approach allows silencing defense systems of choice. We show how this approach enables programming of “untransformable” bacteria, and how it can enhance phage therapy applications
Congrats Jeremy Garb!
tinyurl.com/Syttt
🧵

Our approach allows silencing defense systems of choice. We show how this approach enables programming of “untransformable” bacteria, and how it can enhance phage therapy applications
Congrats Jeremy Garb!
tinyurl.com/Syttt
🧵
www.nature.com/articles/s41...
@natbiotech.nature.com @zhangf.bsky.social

www.nature.com/articles/s41...
@natbiotech.nature.com @zhangf.bsky.social


#BenhurLee #JesseBloom #LisaWagner @wchnicholas.bsky.social and #IleanaCristea. Safe travel everyone! #LoveVirology

#BenhurLee #JesseBloom #LisaWagner @wchnicholas.bsky.social and #IleanaCristea. Safe travel everyone! #LoveVirology

Her lab uncovers the inner workings of viruses and their interaction with the immune system to prevent current and future viral diseases: bit.ly/44peaBa

Her lab uncovers the inner workings of viruses and their interaction with the immune system to prevent current and future viral diseases: bit.ly/44peaBa

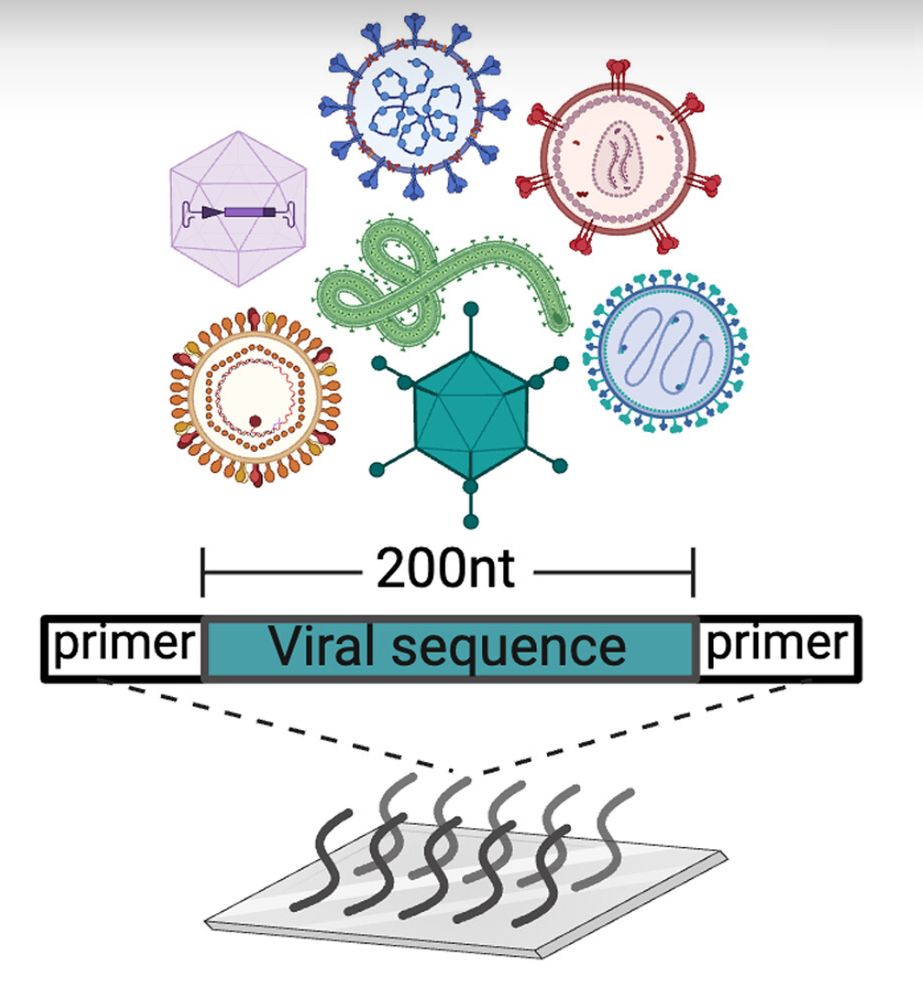
We're excited to share our new publication developing Massively Parallel Ribosome Profiling (MPRP), which uncovered ~4,000 hidden proteins in ~700 viral genomes. www.science.org/doi/10.1126/...

We're excited to share our new publication developing Massively Parallel Ribosome Profiling (MPRP), which uncovered ~4,000 hidden proteins in ~700 viral genomes. www.science.org/doi/10.1126/...