
Functional genomics, Proteostasis, Neurodegeneration
www.shalemlab.org
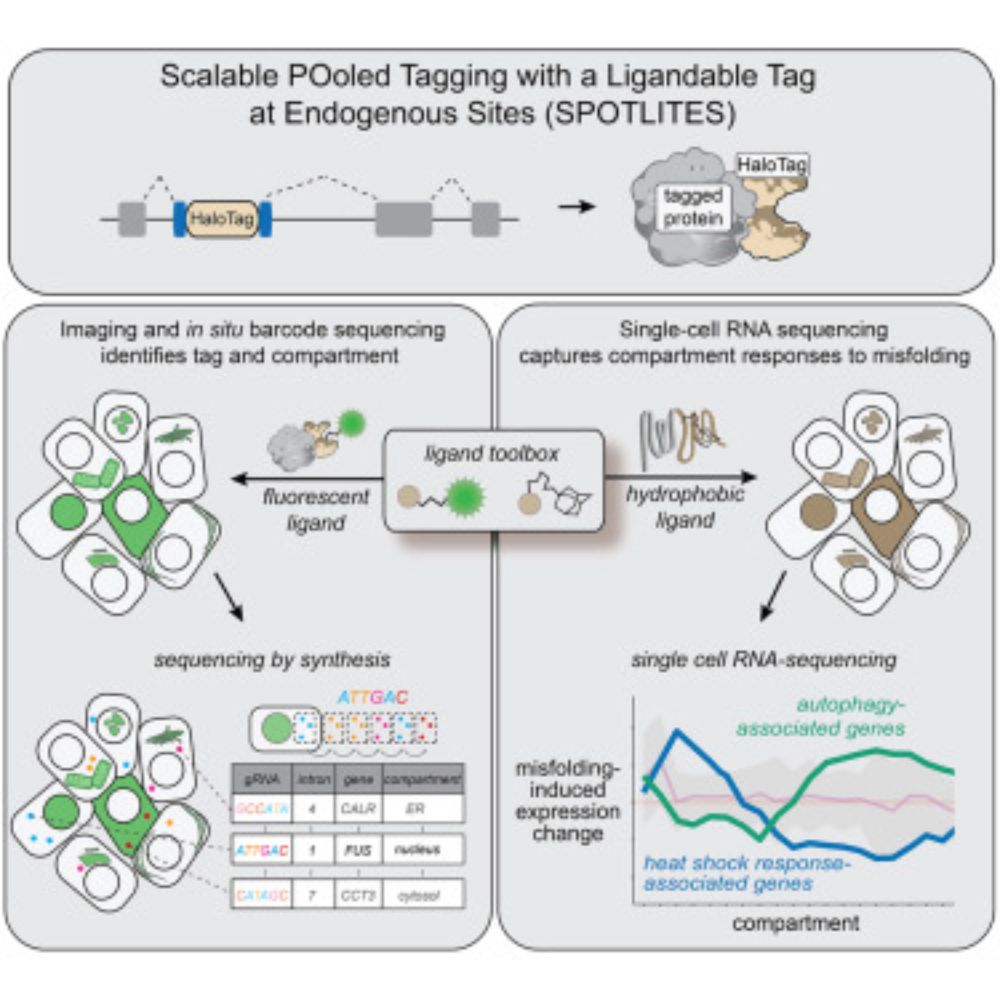
www.cell.com/molecular-ce...
We're excited to share our new publication developing Massively Parallel Ribosome Profiling (MPRP), which uncovered ~4,000 hidden proteins in ~700 viral genomes. www.science.org/doi/10.1126/...

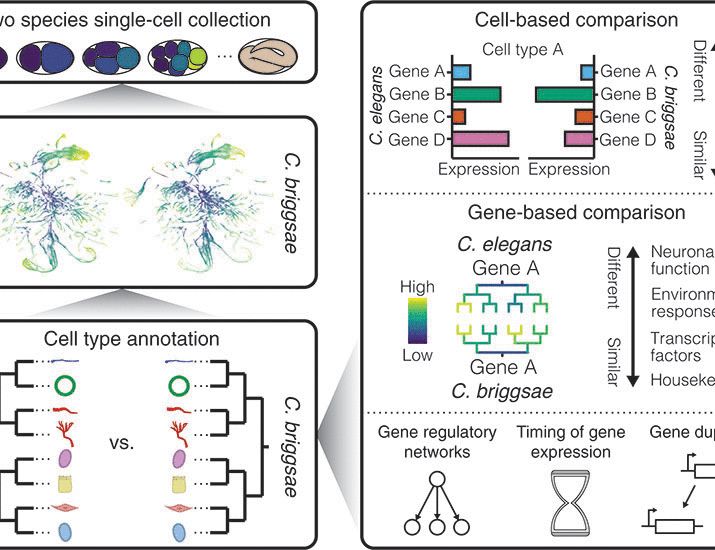
www.cell.com/molecular-ce...

www.cell.com/molecular-ce...
Documentation here!
docs.nimbusimage.com

Documentation here!
docs.nimbusimage.com
One with @dremilygoldberg.bsky.social on lysine lactylation
www.biorxiv.org/content/10.1...
Another with @taabaman.bsky.social on new peptide tools to study caspases www.biorxiv.org/content/10.1...

One with @dremilygoldberg.bsky.social on lysine lactylation
www.biorxiv.org/content/10.1...
Another with @taabaman.bsky.social on new peptide tools to study caspases www.biorxiv.org/content/10.1...
Unfortunately due to the funding source, this position is only available to US citizens or permanent residents.
chemistryjobs.acs.org/job/postdoct...
Unfortunately due to the funding source, this position is only available to US citizens or permanent residents.
chemistryjobs.acs.org/job/postdoct...

careers.chop.edu/us/en/event/...

careers.chop.edu/us/en/event/...
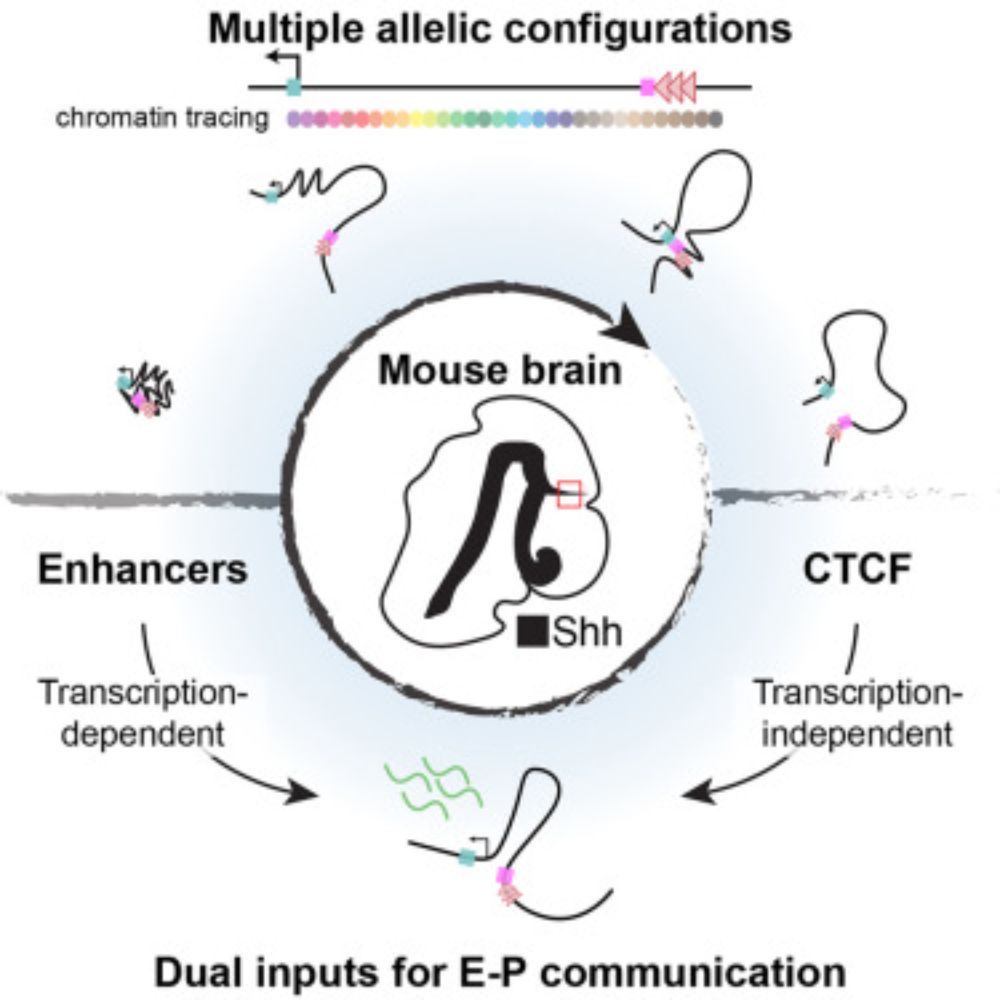
www.nature.com/articles/s41...
www.nature.com/articles/s41...
t.co/wXqtte3pqp
t.co/wXqtte3pqp
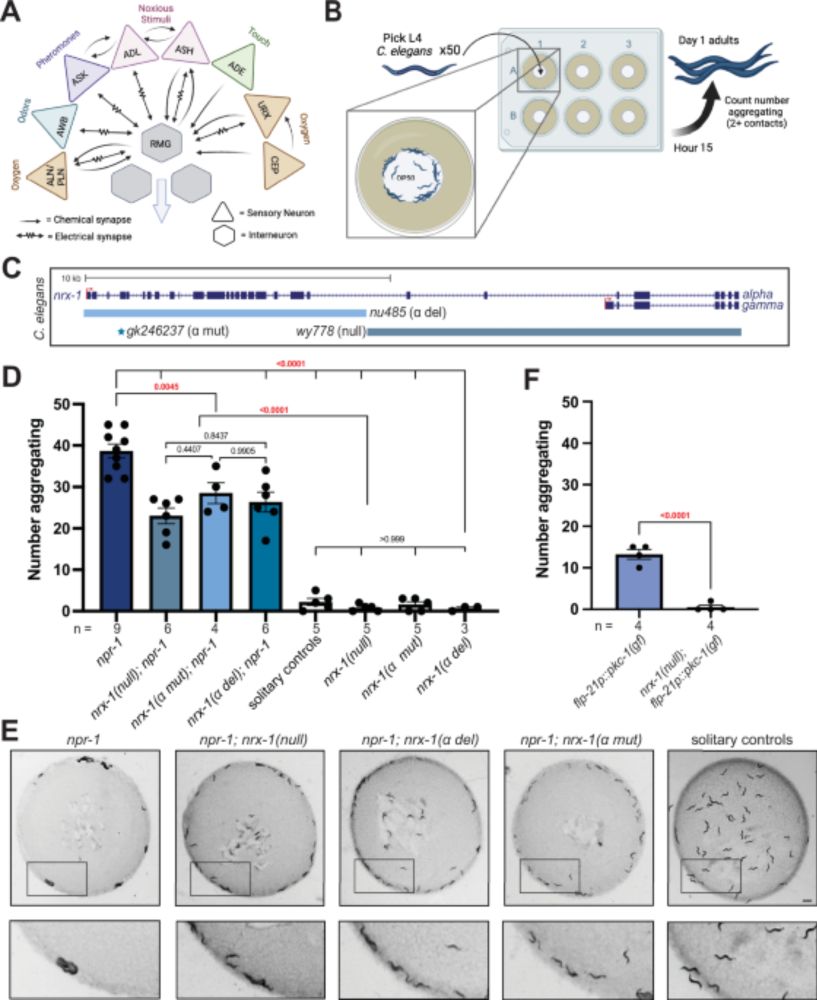
www.biorxiv.org/content/10.1...
A thread...

www.biorxiv.org/content/10.1...
A thread...
www.science.org/doi/10.1126/...
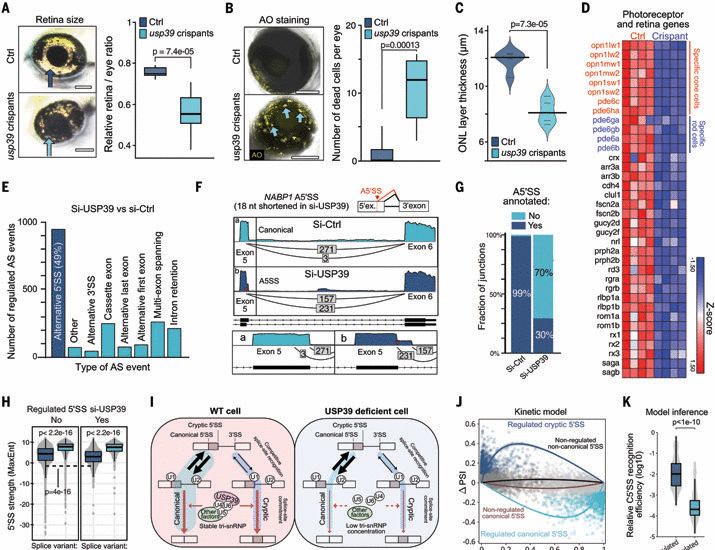
www.science.org/doi/10.1126/...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

