
I'm a PhD student working on ML methods for Drug Discovery at Edinburgh with @ppxasjsm.bsky.social. Looking forward to connecting with fellow ML, CompChem, Bio and Drug Discovery folks!🧬💊💻
Sharing starter packs in 🧵 below that I found helpful in finding people working in the domain.
🔗 www.findaphd.com/phds/project...
#PhD #MachineLearning #DrugDiscovery

🔗 www.findaphd.com/phds/project...
#PhD #MachineLearning #DrugDiscovery




Leveraging all the things we've recently learned training AI models for 1000x impact in science
and this will need data!
More details: huggingface.co/blog/lemater...
Thread:
...

Leveraging all the things we've recently learned training AI models for 1000x impact in science
and this will need data!
More details: huggingface.co/blog/lemater...
Thread:
...
Take a look at our paper in @cellpress.bsky.social!
www.cell.com/cell/fulltex...
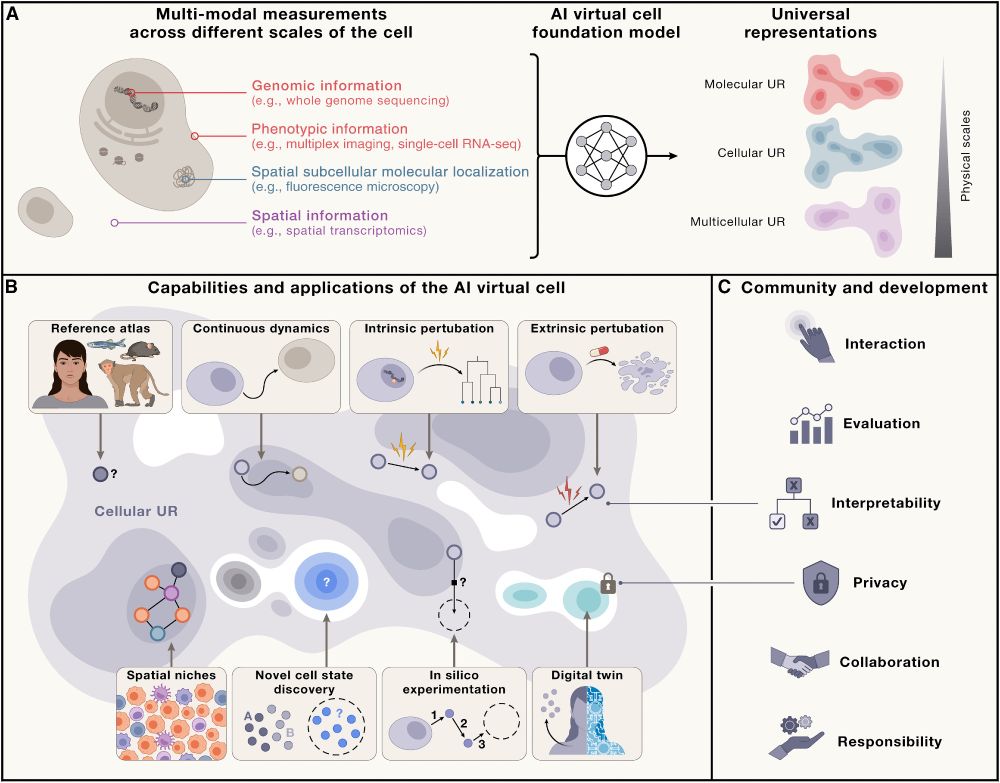
Take a look at our paper in @cellpress.bsky.social!
www.cell.com/cell/fulltex...

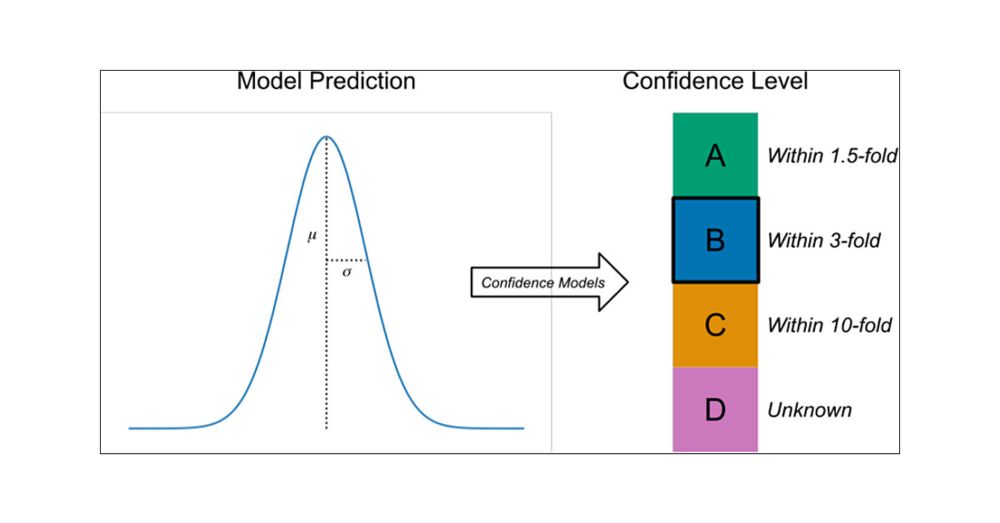
(E.g., where does that “log” come from? Are there other possible formulas?)
Yet there's an intuitive & almost inevitable way to arrive at this expression.
(E.g., where does that “log” come from? Are there other possible formulas?)
Yet there's an intuitive & almost inevitable way to arrive at this expression.
www.nature.com/articles/s43...
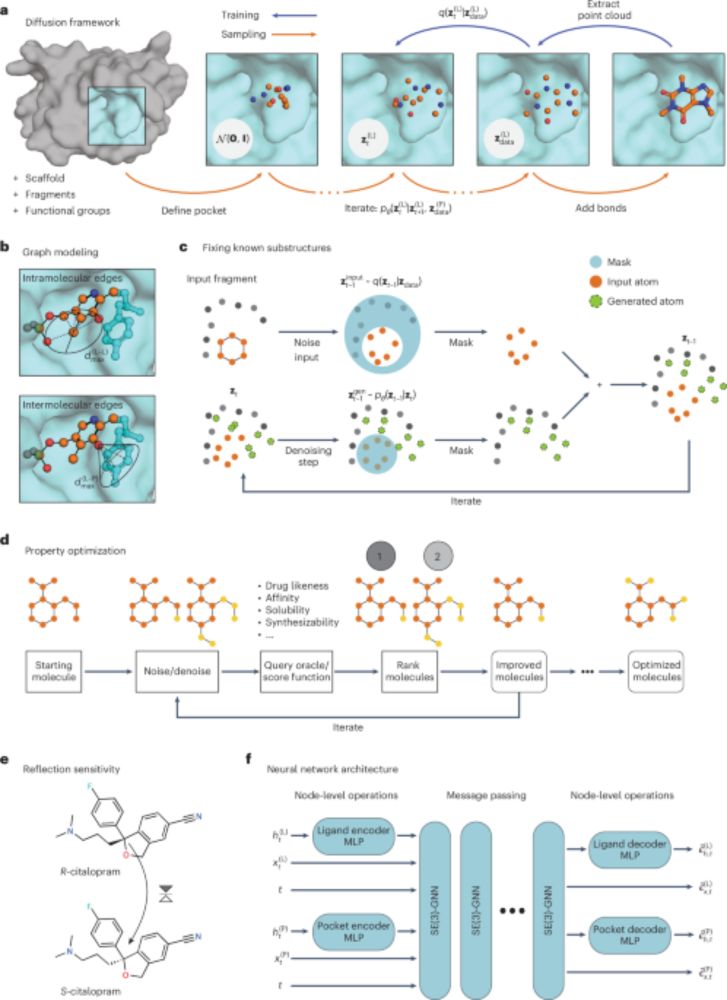
www.nature.com/articles/s43...
Really proud of @amyxlu.bsky.social 's effort leading this project end-to-end!

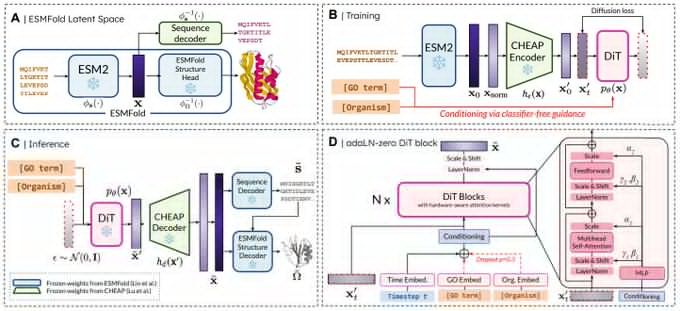

Really proud of @amyxlu.bsky.social 's effort leading this project end-to-end!

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Here's what I found useful - would love to hear your favourites!
🧵1/4
#Science #AcademicSky #PhDSky
Here's what I found useful - would love to hear your favourites!
🧵1/4
#Science #AcademicSky #PhDSky
Outperforms all models at similar GPU RAM usage and tokens throughputs
Blog post: huggingface.co/blog/smolvlm

Outperforms all models at similar GPU RAM usage and tokens throughputs
Blog post: huggingface.co/blog/smolvlm

I'm a PhD student working on ML methods for Drug Discovery at Edinburgh with @ppxasjsm.bsky.social. Looking forward to connecting with fellow ML, CompChem, Bio and Drug Discovery folks!🧬💊💻
Sharing starter packs in 🧵 below that I found helpful in finding people working in the domain.
I'm a PhD student working on ML methods for Drug Discovery at Edinburgh with @ppxasjsm.bsky.social. Looking forward to connecting with fellow ML, CompChem, Bio and Drug Discovery folks!🧬💊💻
Sharing starter packs in 🧵 below that I found helpful in finding people working in the domain.
chemrxiv.org/engage/chemr...
#computationalchemistry #medchem #compchem

chemrxiv.org/engage/chemr...
#computationalchemistry #medchem #compchem
Publication of 10 new foundation models of Proteins, DNA, RNA, methylation, cells, and interactions, evolution, and design in the past couple of weeks!
Unprecedented progress, reviewed in the new Ground Truths
erictopol.substack.com/p/learning-t...

Publication of 10 new foundation models of Proteins, DNA, RNA, methylation, cells, and interactions, evolution, and design in the past couple of weeks!
Unprecedented progress, reviewed in the new Ground Truths
erictopol.substack.com/p/learning-t...
Release Details: github.com/rdk/p2rank/r...
💡 Feedback welcome!
#Bioinformatics #DrugDiscovery
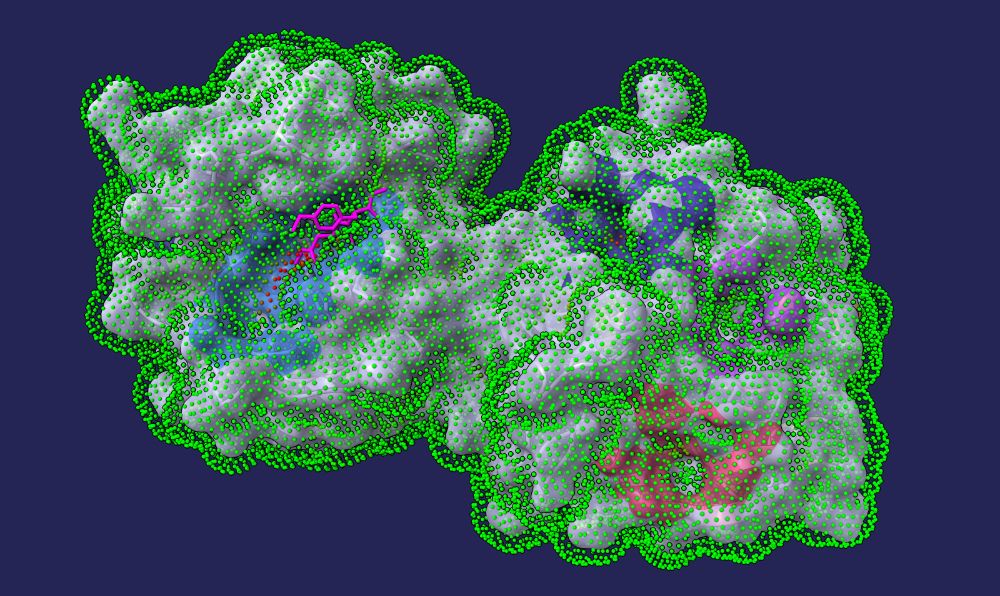
Release Details: github.com/rdk/p2rank/r...
💡 Feedback welcome!
#Bioinformatics #DrugDiscovery
#cancerresearch #bloodflowmodel #HPC #bioengineering
#cancerresearch #bloodflowmodel #HPC #bioengineering
I have compiled a list of now 100+ companies in the 'TechBio' space into a fully open database for the community.
Find here and please share if you like
open.substack.com/pub/harrisbi...

I have compiled a list of now 100+ companies in the 'TechBio' space into a fully open database for the community.
Find here and please share if you like
open.substack.com/pub/harrisbi...

We have three models released based on SMILES, SELFIES and molecular graphs.
More to come shortly - we aim to have a unified collection of state-of art models across all modalities.

We have three models released based on SMILES, SELFIES and molecular graphs.
More to come shortly - we aim to have a unified collection of state-of art models across all modalities.

