
🔬 @iocbprague.bsky.social, prev. PhD @cusbg.bsky.social @mff.unikarlova.cuni.cz
(1/n)




A #research article about their vast #massspec library has been published in @natmethods.nature.com with Corinna Brungs & @robinschmidt.bsky.social as the first two authors.
See more ► www.uochb.cz/en/news/732

Join us for a special anniversary program 15–21 Sept 2025. 🎉
@unikarlova.cuni.cz @botanickazahradauk.bsky.social




Join us for a special anniversary program 15–21 Sept 2025. 🎉
@unikarlova.cuni.cz @botanickazahradauk.bsky.social
@mdc-biocat.bsky.social - how did comparison of Boltz-2 to PocketVina turn out?
@mdc-biocat.bsky.social - how did comparison of Boltz-2 to PocketVina turn out?

#CECAM @cecamevents.bsky.social @iocbprague.bsky.social

#CECAM @cecamevents.bsky.social @iocbprague.bsky.social
Talks by Ulf Ryde, Ran Friedman & Charles-Alexandre Mattelaer explored QM methods for binding affinity, energy decomposition & binding geometries. Great discussions on rigorous QM in drug design!
#compchem @iocbprague.bsky.social



Talks by Ulf Ryde, Ran Friedman & Charles-Alexandre Mattelaer explored QM methods for binding affinity, energy decomposition & binding geometries. Great discussions on rigorous QM in drug design!
#compchem @iocbprague.bsky.social
@iocbprague.bsky.social @cecamevents.bsky.social



@iocbprague.bsky.social @cecamevents.bsky.social
M.Lepsik on #SQM scoring on AI-generated structures,M.Hennemann on local properties at P-L interfaces & I.Gordiy on enhanced sampling with ML potentials.Inspiring end before our social dinner!
@cecamevents.bsky.social @iocbprague.bsky.social @iocbtech.bsky.social



M.Lepsik on #SQM scoring on AI-generated structures,M.Hennemann on local properties at P-L interfaces & I.Gordiy on enhanced sampling with ML potentials.Inspiring end before our social dinner!
@cecamevents.bsky.social @iocbprague.bsky.social @iocbtech.bsky.social
#CECAMinPrague @iocbprague.bsky.social @cecamevents.bsky.social @iocbtech.bsky.social

#CECAMinPrague @iocbprague.bsky.social @cecamevents.bsky.social @iocbtech.bsky.social
Talks by Kennie Merz, Jan Řezáč, Markus Hermann & Hamza Ibrahim sparked great discussion on #QM, #ML & #DrugDesign
Thanks to @iocbprague.bsky.social, @iocbtech.bsky.social & @cecamevents.bsky.social for support!
#compchem #SQM



Talks by Kennie Merz, Jan Řezáč, Markus Hermann & Hamza Ibrahim sparked great discussion on #QM, #ML & #DrugDesign
Thanks to @iocbprague.bsky.social, @iocbtech.bsky.social & @cecamevents.bsky.social for support!
#compchem #SQM
#compchem #SQM @iocbtech.bsky.social @cecamevents.bsky.social @iocbprague.bsky.social

#compchem #SQM @iocbtech.bsky.social @cecamevents.bsky.social @iocbprague.bsky.social
They celebrate with a Symposium on September 16th! I'll be presenting our work: "An Atlas of Energetic Evolutionary Constraints Across Protein Families"
Unbelievable selection of speakers! So if you are around London, you can register to attend:
tinyurl.com/2pnk8hj5


They celebrate with a Symposium on September 16th! I'll be presenting our work: "An Atlas of Energetic Evolutionary Constraints Across Protein Families"
Unbelievable selection of speakers! So if you are around London, you can register to attend:
tinyurl.com/2pnk8hj5
www.nature.com/articles/s41...
www.nature.com/articles/s41...





🧪
(Includes some #bayesian #rstats inspired by @rmcelreath.bsky.social)

🧪
(Includes some #bayesian #rstats inspired by @rmcelreath.bsky.social)
patwalters.github.io/Three-Papers...

patwalters.github.io/Three-Papers...
![Travis Kalanick, the founder of Uber who no longer works at the company, appeared on All-In to talk with hosts Jason Calacanis and Chamath Palihapitiya about the future of technology. When the topic turned to AI, Kalanick discussed how he uses xAI’s Grok, which went haywire last week, praising Adolf Hitler and advocating for a second Holocaust against Jews.
“I’ll go down this thread with [Chat]GPT or Grok and I’ll start to get to the edge of what’s known in quantum physics and then I’m doing the equivalent of vibe coding, except it’s vibe physics,” Kalanick explained. “And we’re approaching what’s known. And I’m trying to poke and see if there’s breakthroughs to be had. And I’ve gotten pretty damn close to some interesting breakthroughs just doing that.”](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:qlnirjwff4ibiw3wchmleszo/bafkreiac2szurc5dnw6klquhpjmo7iafg4n6fwmrqyred7jrrph6h6mptq@jpeg)
The title is "Mutations within the predicted fragment-binding region of FAM83G/SACK1G abolish ints interaction with the Ser/Thr kinase CK1α".
🔗 Pre-print: www.biorxiv.org/content/10.1...
The title is "Mutations within the predicted fragment-binding region of FAM83G/SACK1G abolish ints interaction with the Ser/Thr kinase CK1α".
🔗 Pre-print: www.biorxiv.org/content/10.1...
github.com/martinpacesa...
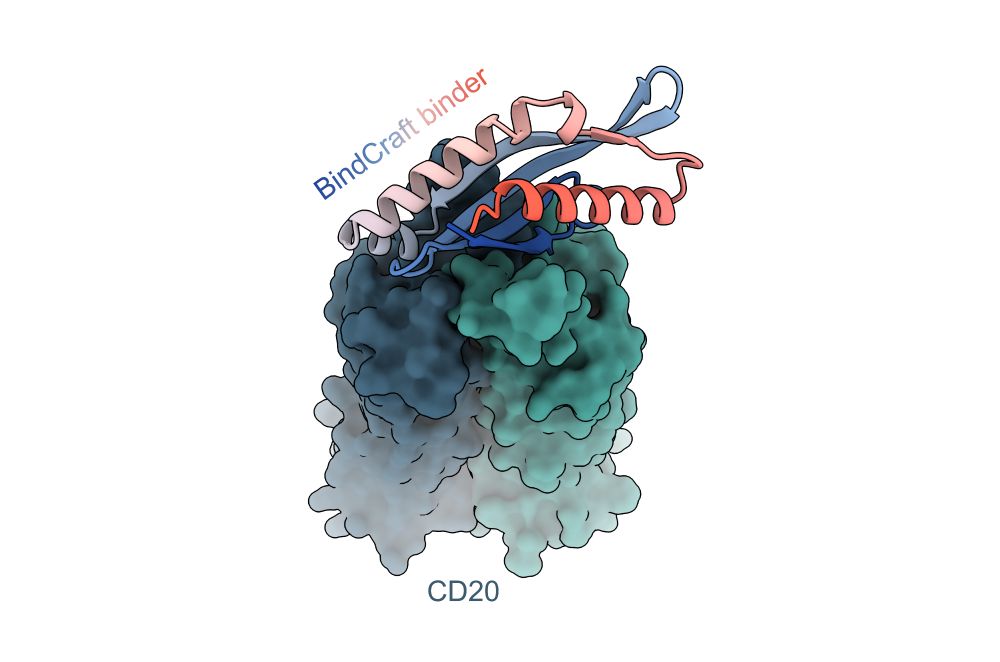
github.com/martinpacesa...


