We had a great 3DSIG/3DBioinfo event in "sunny" Barcelona this week!! I have enjoyed so much to help organising it & to have such great minds all together sharing science for 3 days!!
Aufwiedersehen friends! For more science & friendships to come! ⭐




www.biorxiv.org/content/10.1...
🧵👇 (1/n)

Independent Group Leader Positions in Computational Biology @humantechnopole.bsky.social!
Are you ready to start your own lab? Do you know someone who is? Repost this + share with everyone who might want to know about it. Thanks!!! 🙏
More details below... check it out! 🧵 1/3

Independent Group Leader Positions in Computational Biology @humantechnopole.bsky.social!
Are you ready to start your own lab? Do you know someone who is? Repost this + share with everyone who might want to know about it. Thanks!!! 🙏
More details below... check it out! 🧵 1/3



Full update below.

Full update below.
@mattbashton.bsky.social and Ricardo Rajsbaum is out in BioRxiv. www.biorxiv.org/content/10.1...

@mattbashton.bsky.social and Ricardo Rajsbaum is out in BioRxiv. www.biorxiv.org/content/10.1...


https://buff.ly/3ATRAFT

The ISCB 3DSig together with the 3DBioinfo @ELIXIREurope community we are organising a joint conference in Barcelona March 19-21, 2025!
If you work in Structural Bioinformatics or Computational Biophysics communities register ASAP! Only 120 spots!
www.iscb.org/3dbioinfo202...

The ISCB 3DSig together with the 3DBioinfo @ELIXIREurope community we are organising a joint conference in Barcelona March 19-21, 2025!
If you work in Structural Bioinformatics or Computational Biophysics communities register ASAP! Only 120 spots!
www.iscb.org/3dbioinfo202...


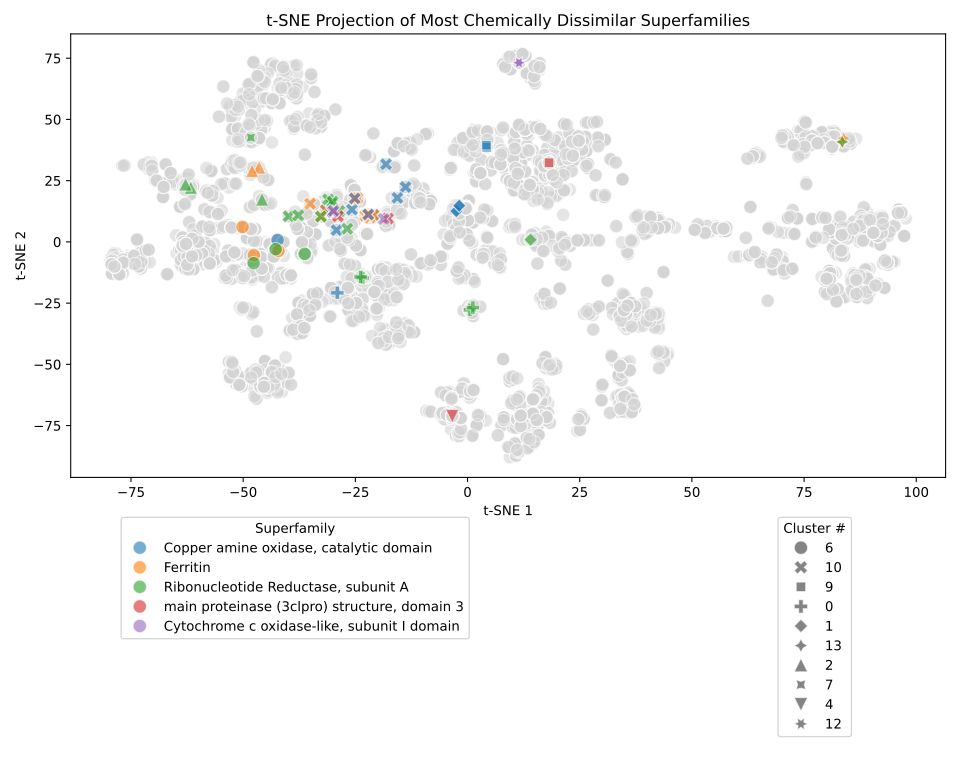
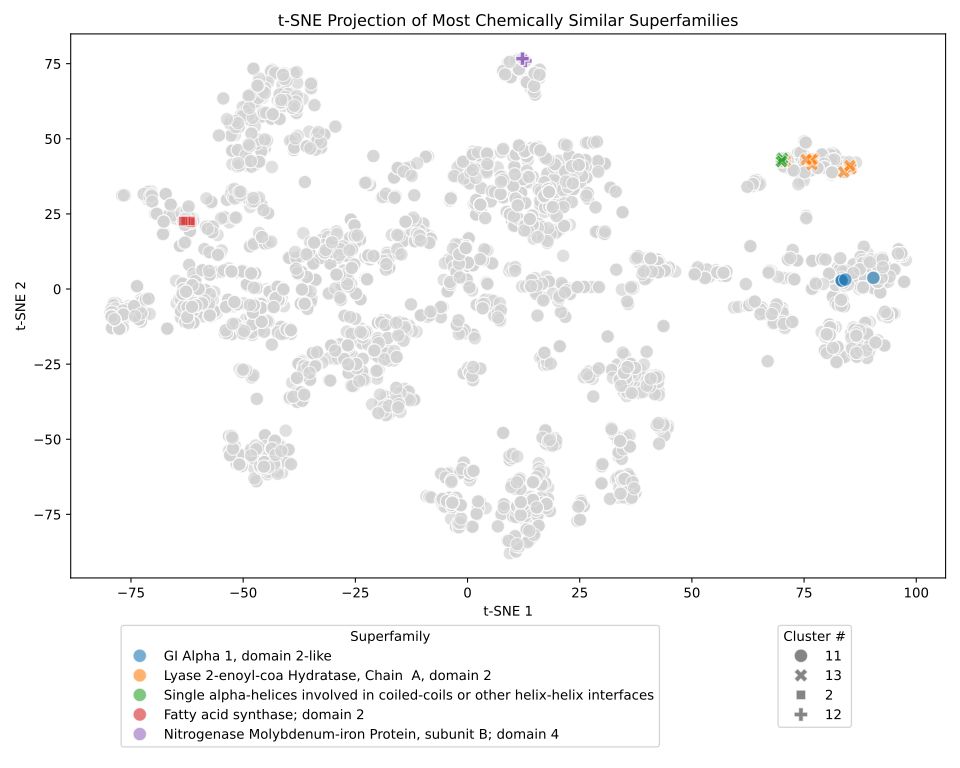
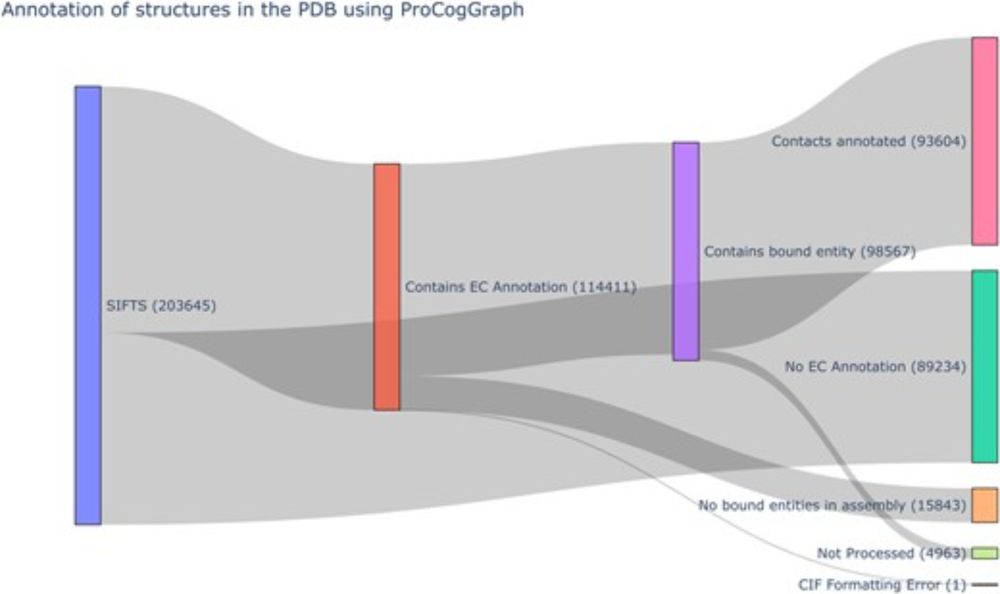
www.thelancet.com/journals/lan...

www.thelancet.com/journals/lan...


