
Structural and High Throughout Sequencing Bioinformatics, Protein Domains, Pandemic Preparedness.
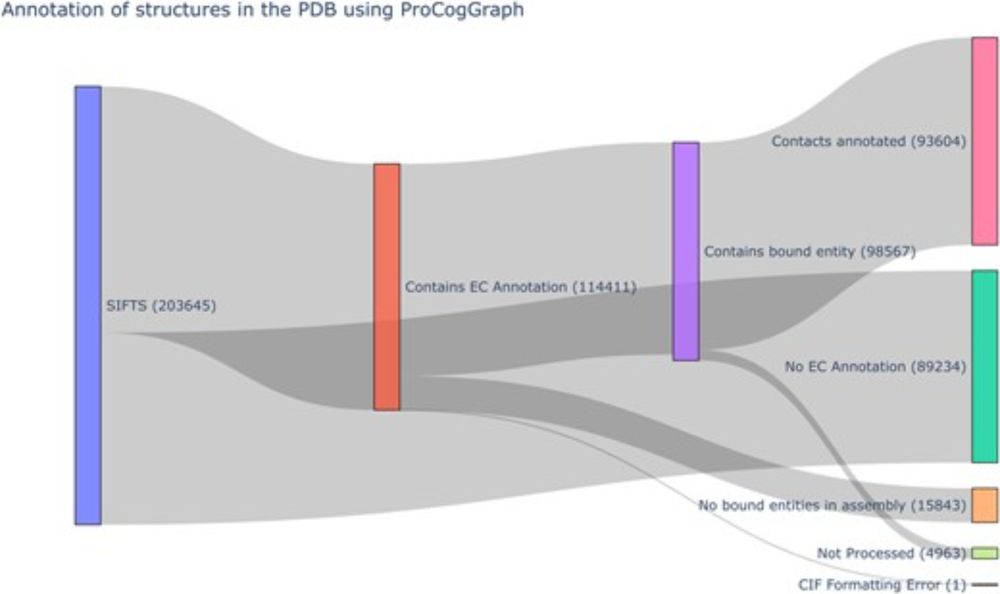
📄 www.biorxiv.org/content/10.1...
💾 github.com/steineggerla...
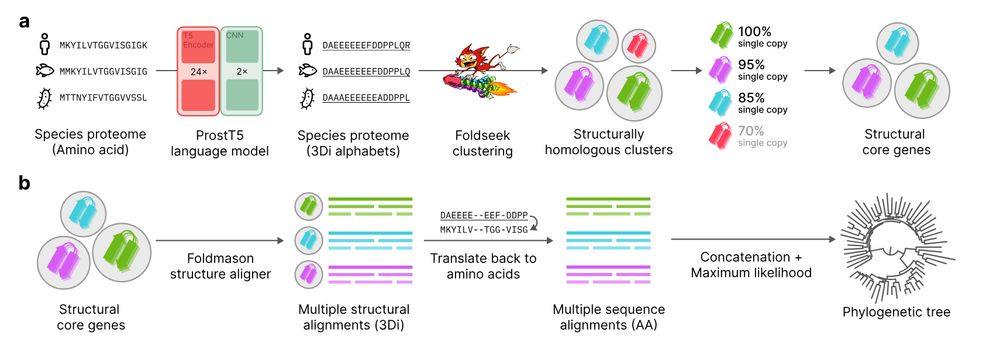
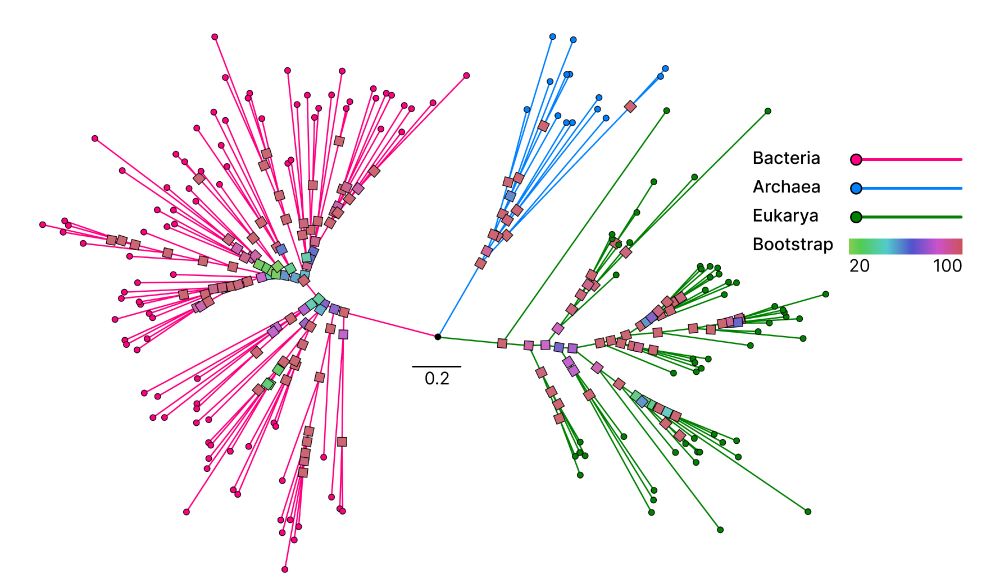
📄 www.biorxiv.org/content/10.1...
💾 github.com/steineggerla...
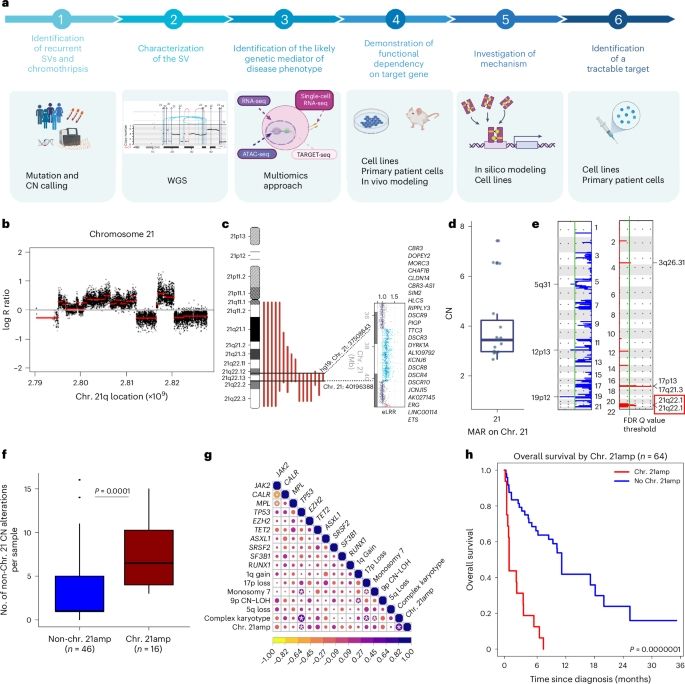
Read the full discussion here: https://doi.org/10.1093/bioadv/vbaf044
#DataIntegrity #GenerativeAI




www.northumbria.ac.uk/about-us/new...

www.northumbria.ac.uk/about-us/new...




@mattbashton.bsky.social and Ricardo Rajsbaum is out in BioRxiv. www.biorxiv.org/content/10.1...

@mattbashton.bsky.social and Ricardo Rajsbaum is out in BioRxiv. www.biorxiv.org/content/10.1...



Details here:
www.findaphd.com/phds/project...

Details here:
www.findaphd.com/phds/project...

Welsh speakers: I apologise for my pronunciation of Llanfairpwllgwyngyllgogerychwyrndrobwllllantysiliogogogoch


www.npr.org/2024/10/22/n...
"LA is seeing cases of dengue, the range of which may be growing due to climate change"
www.npr.org/2024/10/22/n...
"LA is seeing cases of dengue, the range of which may be growing due to climate change"
This commentary, led by Anthony Gitter, discusses the importance of data, code, and model sharing for further scientific progress and independent evaluation of claims in biomedical #AI 🧪🧬🖥️ #AcademicSky
This commentary, led by Anthony Gitter, discusses the importance of data, code, and model sharing for further scientific progress and independent evaluation of claims in biomedical #AI 🧪🧬🖥️ #AcademicSky








