
Open access link: www.nature.com/articles/s41...
(1/9)
Open access link: www.nature.com/articles/s41...
(1/9)


www.biorxiv.org/content/10.6...

www.biorxiv.org/content/10.6...
It’s a great read on the impact of DNA secondary structures on eukaryotic replication fork progression - a totally unbiased opinion, of course!
www.sciencedirect.com/science/arti...
@adityasethi.bsky.social @billiedelpino.bsky.social

It’s a great read on the impact of DNA secondary structures on eukaryotic replication fork progression - a totally unbiased opinion, of course!
www.sciencedirect.com/science/arti...
@adityasethi.bsky.social @billiedelpino.bsky.social
www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...
@penengolab.bsky.social and collaborators
www.embopress.org/doi/full/10....


Niels Mailand, Robert Shearer et al profile a panel of #ubiquitin replacement cell lines, implicating K29 chains in chromatin regulation via SUV39H1 destabilization
www.embopress.org/doi/full/10....

Niels Mailand, Robert Shearer et al profile a panel of #ubiquitin replacement cell lines, implicating K29 chains in chromatin regulation via SUV39H1 destabilization
www.embopress.org/doi/full/10....
www.nature.com/articles/s41...

www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

www.nature.com/articles/s41...
www.nature.com/articles/s41...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
Here, we reveal the unique, molecular mechanism by which USP1/UAF1 cleaves ubiquitin chains on PCNA, which may direct DNA damage tolerance.
doi.org/10.1038/s414...
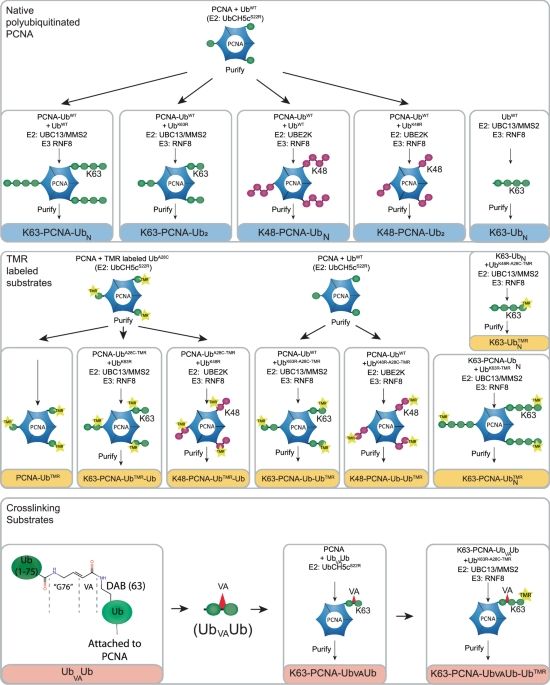
Here, we reveal the unique, molecular mechanism by which USP1/UAF1 cleaves ubiquitin chains on PCNA, which may direct DNA damage tolerance.
doi.org/10.1038/s414...
uobevents.eventsair.com/birmingham-c...
uobevents.eventsair.com/birmingham-c...
Please get in touch with your CV if you are interested; closing date: 27th June 2025.
More details here: candidate.hr-manager.net/ApplicationI...
Please get in touch with your CV if you are interested; closing date: 27th June 2025.
More details here: candidate.hr-manager.net/ApplicationI...


www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

