Chromatin, Hi-C, gene regulation
Co-organizer of Sci.STEPS mentoring program
compartment where extensive loop
extrusion isolates domains from
other such domains
Hi-C analysis of silkworm chromosomes by Drinnenberg, Muller, Mirny et al reveals new combination of these mechanisms, and a new, secluded “S” compartment
link.springer.com/article/10.1...

compartment where extensive loop
extrusion isolates domains from
other such domains
doi.org/10.1093/nar/...

doi.org/10.1093/nar/...
Deadline 15th December 2025
#epigenetics Please share 🙏
Deadline 15th December 2025
#epigenetics Please share 🙏

I am delighted that our paper exploring the impact of Neanderthal-derived variants on the activity of a disease-associated craniofacial enhancer has been published in Development today!
journals.biologists.com/dev/article/...

I am delighted that our paper exploring the impact of Neanderthal-derived variants on the activity of a disease-associated craniofacial enhancer has been published in Development today!
journals.biologists.com/dev/article/...
We invite #mentors from varied backgrounds and stages of their #scientific careers to apply and share their knowledge and expertise with our #mentees.
✏ Register here: lnkd.in/edZggY32
🗓 Deadline: November 10th 2025
👀more about #SciSTEPS: www.scisteps.org

We invite #mentors from varied backgrounds and stages of their #scientific careers to apply and share their knowledge and expertise with our #mentees.
✏ Register here: lnkd.in/edZggY32
🗓 Deadline: November 10th 2025
👀more about #SciSTEPS: www.scisteps.org

www.biorxiv.org/content/10.1...
A🧵👇
www.biorxiv.org/content/10.1...
A🧵👇
www.biorxiv.org/content/10.1...
A🧵👇
We find the rate of cohesin loop extrusion in cells is set by NIPBL dosage and tunes many aspects of chromosome folding.
This provides a molecular basis for NIPBL haploinsufficiency in humans. 🧵👇
www.biorxiv.org/content/10.1...

We find the rate of cohesin loop extrusion in cells is set by NIPBL dosage and tunes many aspects of chromosome folding.
This provides a molecular basis for NIPBL haploinsufficiency in humans. 🧵👇
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
1/
www.biorxiv.org/content/10.1...
1/
www.biorxiv.org/content/10.1...
How does enhancer location within a TAD control transcriptional bursts from a cognate promoter?
Experiments by Jana Tünnermann and modelling by Gregory Roth

that explores how PRC1-mediated H2AK119ub deposition safeguards lineage fidelity during forebrain development. Fantastic work by @lucyadoyle.bsky.social and our colleagues & collaborators. 🥳🧬🧠🔬
www.biorxiv.org/content/10.1...

that explores how PRC1-mediated H2AK119ub deposition safeguards lineage fidelity during forebrain development. Fantastic work by @lucyadoyle.bsky.social and our colleagues & collaborators. 🥳🧬🧠🔬
www.biorxiv.org/content/10.1...
Deadline July 15th.
www.mdc-berlin.de/career/jobs/...
Deadline July 15th.
www.mdc-berlin.de/career/jobs/...
Learn more at: oxbow.readthedocs.io
Learn more at: oxbow.readthedocs.io
www.fmi.ch/education-ca...
www.fmi.ch/education-ca...
Come join our HFSP team to uncover how chromatin moves in cells and what this means for genome function! Great opportunity to combine single-cell genomics, live imaging and polymer physics in the unique mammalian retina with @andersshansen.bsky.social, Davide Michieletto & Sandra Tenreiro

Come join our HFSP team to uncover how chromatin moves in cells and what this means for genome function! Great opportunity to combine single-cell genomics, live imaging and polymer physics in the unique mammalian retina with @andersshansen.bsky.social, Davide Michieletto & Sandra Tenreiro
Come join my lab @Novartis Oncology in Basel 🇨🇭, and employ genome engineering and functional genomics approaches to study how oncogenic Transcription Factors function at the molecular level (and how to drug them)
3+1 years! Apply!
Please RP
www.novartis.com/careers/care...

Come join my lab @Novartis Oncology in Basel 🇨🇭, and employ genome engineering and functional genomics approaches to study how oncogenic Transcription Factors function at the molecular level (and how to drug them)
3+1 years! Apply!
Please RP
www.novartis.com/careers/care...
pubmed.ncbi.nlm.nih.gov/40436628/
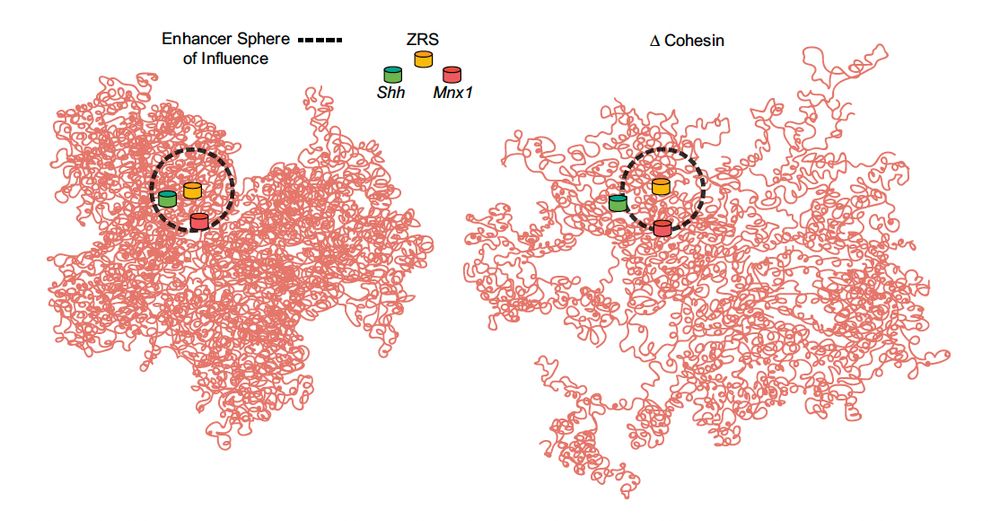
pubmed.ncbi.nlm.nih.gov/40436628/
www.findaphd.com/phds/project...
Excited to collaborate with @hannahlong.bsky.social and Daria Bunina (@uoe-igc.bsky.social/@mdc-berlin.bsky.social).
Please share 🙏
#epigenetics

www.findaphd.com/phds/project...
Excited to collaborate with @hannahlong.bsky.social and Daria Bunina (@uoe-igc.bsky.social/@mdc-berlin.bsky.social).
Please share 🙏
#epigenetics
🧫 Are you interested in mechanisms of gene regulation, human facial development and disease? How our DNA shapes the way we look? Do you love chromatin and enhancers? 🧬
If so, reach out for an informal chat! 💬
longlab.co.uk
🧫 Are you interested in mechanisms of gene regulation, human facial development and disease? How our DNA shapes the way we look? Do you love chromatin and enhancers? 🧬
If so, reach out for an informal chat! 💬
longlab.co.uk
www.biorxiv.org/content/10.1...
How does enhancer location within a TAD control transcriptional bursts from a cognate promoter?
Experiments by Jana Tünnermann and modelling by Gregory Roth

www.biorxiv.org/content/10.1...
How does enhancer location within a TAD control transcriptional bursts from a cognate promoter?
Experiments by Jana Tünnermann and modelling by Gregory Roth

