

318 brand-new protein families + 4 new clans! 🧬
Total collection now: 24,736 entries! 📈
Get exploring via InterPro website, API & Pfam FTP!
💻🧪
Melissa Harrison shares a @pfamdb.bsky.social use case:
Pfam uses AI to extract information about known protein domains
This helps predict protein function & identify potential drug targets! 💊
#ChooseOpenWebinar for live updates 💡
#MedLib #PhDsky #OpenAccess
Melissa Harrison shares a @pfamdb.bsky.social use case:
Pfam uses AI to extract information about known protein domains
This helps predict protein function & identify potential drug targets! 💊
#ChooseOpenWebinar for live updates 💡
#MedLib #PhDsky #OpenAccess
🎤 Multiple talks & 📊 posters on protein analysis & classification
💻🧪



🎤 Multiple talks & 📊 posters on protein analysis & classification
💻🧪
Join us July 20-24 at the world's largest #bioinformatics conference!
#ISMBECCB2025 #ProteinAnalysis #ComputationalBiology
💻🧪

Join us July 20-24 at the world's largest #bioinformatics conference!
#ISMBECCB2025 #ProteinAnalysis #ComputationalBiology
💻🧪
🎤 Talk 08/07 16:50, amphi E: Pfam: embracing AI/ML by @typhainepl.bsky.social
📊 Poster #97: InterPro: Accelerating Protein Annotation with AI by @matthiasblum.bsky.social
💻🧪

Get the answers in a webinar with Creative Commons, #EuropePMC & EMBL
🗓️ 9 Jul
⏰ 15:00 UTC
🔗 embl-org.zoom.us/webinar/regi...
#OpenScience #AcademicSky 🧪

Get the answers in a webinar with Creative Commons, #EuropePMC & EMBL
🗓️ 9 Jul
⏰ 15:00 UTC
🔗 embl-org.zoom.us/webinar/regi...
#OpenScience #AcademicSky 🧪
318 brand-new protein families + 4 new clans! 🧬
Total collection now: 24,736 entries! 📈
Get exploring via InterPro website, API & Pfam FTP!
💻🧪


318 brand-new protein families + 4 new clans! 🧬
Total collection now: 24,736 entries! 📈
Get exploring via InterPro website, API & Pfam FTP!
💻🧪
✨ Updated to Pfam 37.4
✨ 683 new entries added
✨ Enhanced protein classification coverage
Explore the latest annotations and discover new protein insights.
Explore it now: interpro.ebi.ac.uk 💻🧪🧬
#Bioinformatics #ProteinScience #Pfam #EMBLEBI


✨ Updated to Pfam 37.4
✨ 683 new entries added
✨ Enhanced protein classification coverage
Explore the latest annotations and discover new protein insights.
Explore it now: interpro.ebi.ac.uk 💻🧪🧬
#Bioinformatics #ProteinScience #Pfam #EMBLEBI
🧪 Cleaner layout
🧪 Interactive 3D viewers
🧪 Contextual annotations
🧪 Easier navigation
See for yourself:
wwwdev.ebi.ac.uk/pdbe/entry/p...
#PDBeBeta #Bioinformatics #Biochemistry #Science
🧪 Cleaner layout
🧪 Interactive 3D viewers
🧪 Contextual annotations
🧪 Easier navigation
See for yourself:
wwwdev.ebi.ac.uk/pdbe/entry/p...
#PDBeBeta #Bioinformatics #Biochemistry #Science
1.8 billion AI-driven protein annotations developed with Google DeepMind are now live.
✨ Look for the sparkles icon to identify them
📊 Options menu lets you view: InterPro only | InterPro-N-only | Stacked
💻🧪
1.8 billion AI-driven protein annotations developed with Google DeepMind are now live.
✨ Look for the sparkles icon to identify them
📊 Options menu lets you view: InterPro only | InterPro-N-only | Stacked
💻🧪
For the first time, see AlphaFold-derived structural domains directly alongside InterPro annotations in one powerful visualisation. Bridging the sequence-structure divide in protein analysis. 💻🧪
Explore now: www.ebi.ac.uk/interpro/pro...
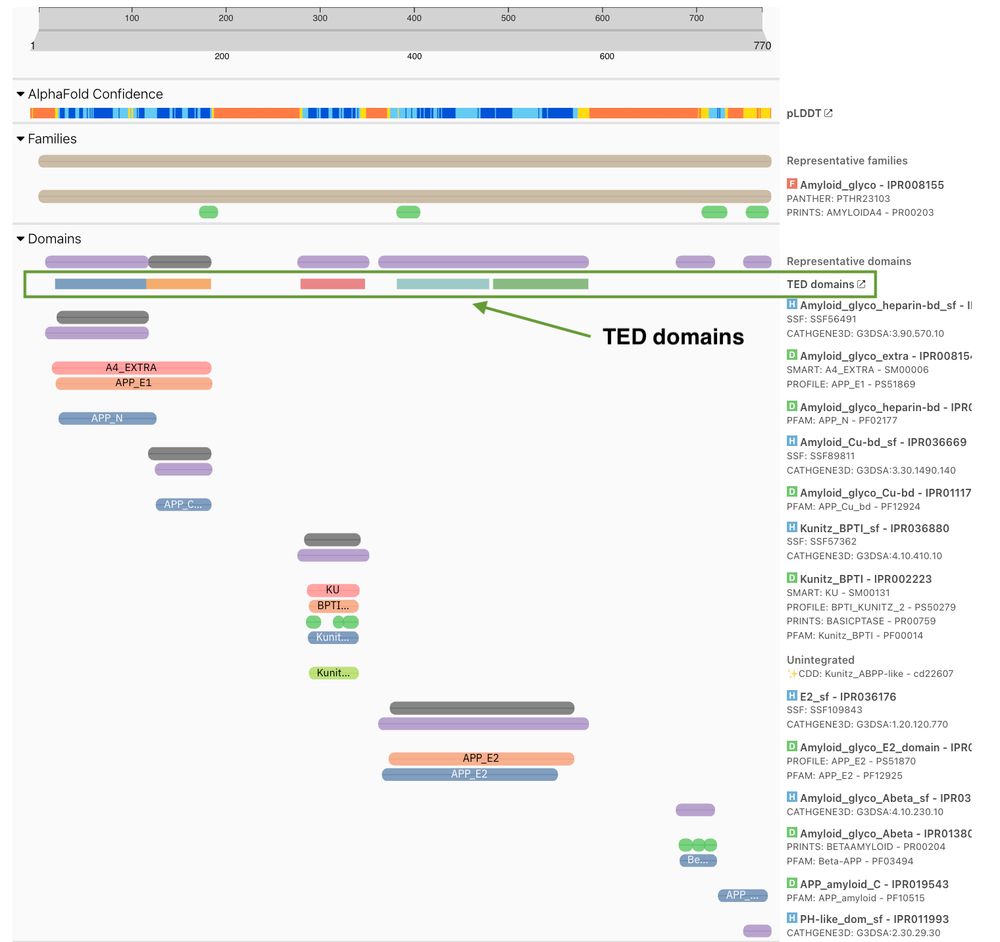
For the first time, see AlphaFold-derived structural domains directly alongside InterPro annotations in one powerful visualisation. Bridging the sequence-structure divide in protein analysis. 💻🧪
Explore now: www.ebi.ac.uk/interpro/pro...
Learn how InterPro-N, TED domains and viral protein structures are transforming protein classification: ebi.ac.uk/about/news/u...
Learn how InterPro-N, TED domains and viral protein structures are transforming protein classification: ebi.ac.uk/about/news/u...
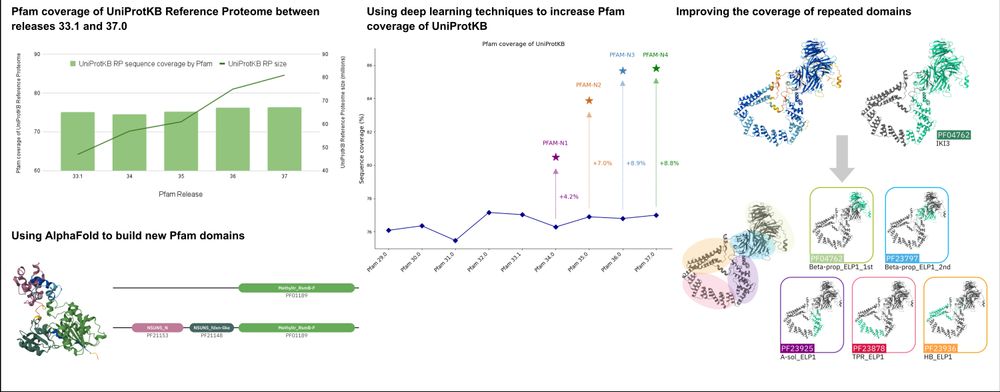
353 incredible new protein families + 7 new clans! 🧬✨
Revolutionise your research NOW via @interprodb.bsky.social website, API & Pfam FTP!

353 incredible new protein families + 7 new clans! 🧬✨
Revolutionise your research NOW via @interprodb.bsky.social website, API & Pfam FTP!
✨Update to Pfam 37.3, PROSITE patterns 2025_01
PROSITE profiles 2025_01 and HAMAP 2025_01
✨342 new entries
✨Addition of InterPro-N annotations
✨Addition of BFVD structure predictions from @martinsteinegger.bsky.social's lab
Have a look: ebi.ac.uk/interpro/htt...
💻🧪🆕




✨Update to Pfam 37.3, PROSITE patterns 2025_01
PROSITE profiles 2025_01 and HAMAP 2025_01
✨342 new entries
✨Addition of InterPro-N annotations
✨Addition of BFVD structure predictions from @martinsteinegger.bsky.social's lab
Have a look: ebi.ac.uk/interpro/htt...
💻🧪🆕
🧶🧬🖥️🧪

✨Update to Pfam 37.2, NCBIfam 17.0, CDD 3.21.
✨661 new entries, including 682 signatures from the Pfam (663), NCBIfam (5), PANTHER (5), CDD (9) databases. Have a look: ebi.ac.uk/interpro/ 💻🧪

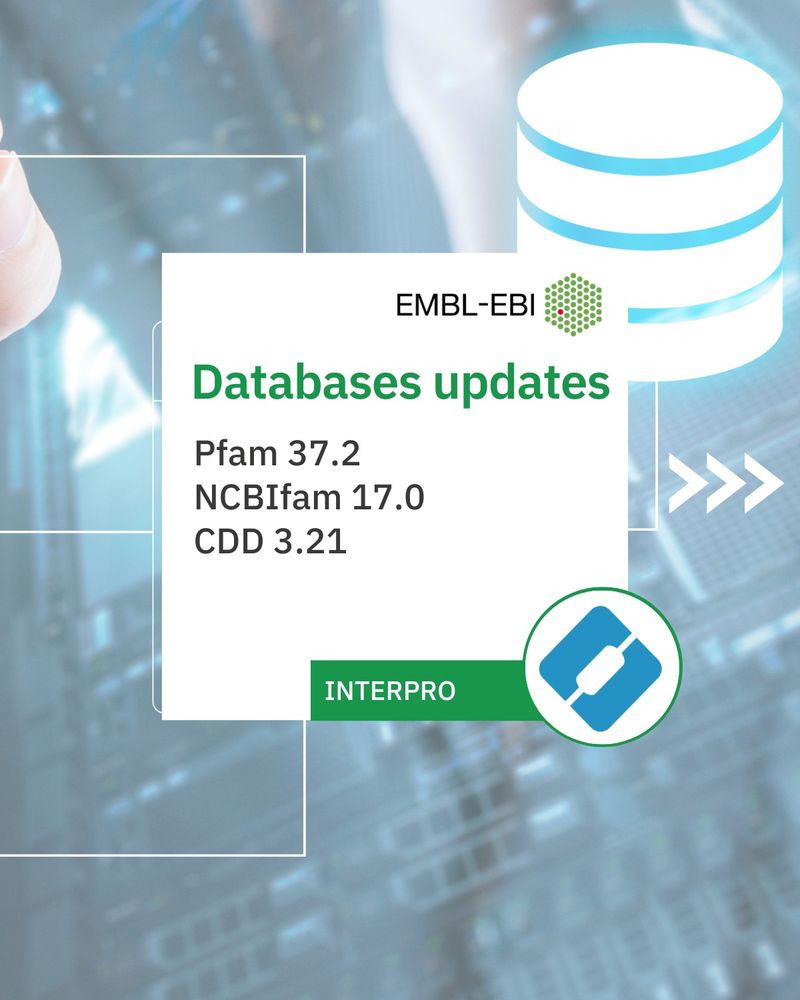


✨Update to Pfam 37.2, NCBIfam 17.0, CDD 3.21.
✨661 new entries, including 682 signatures from the Pfam (663), NCBIfam (5), PANTHER (5), CDD (9) databases. Have a look: ebi.ac.uk/interpro/ 💻🧪

But things get more challenging for protein families.
A new method helps improve low confidence predictions within protein families.
academic.oup.com/bioinformati...
But things get more challenging for protein families.
A new method helps improve low confidence predictions within protein families.
academic.oup.com/bioinformati...
1) Curation data are now in Overview
2) HMM information and logo are now in Profile HMM
Have a look: www.ebi.ac.uk/interpro/ent...
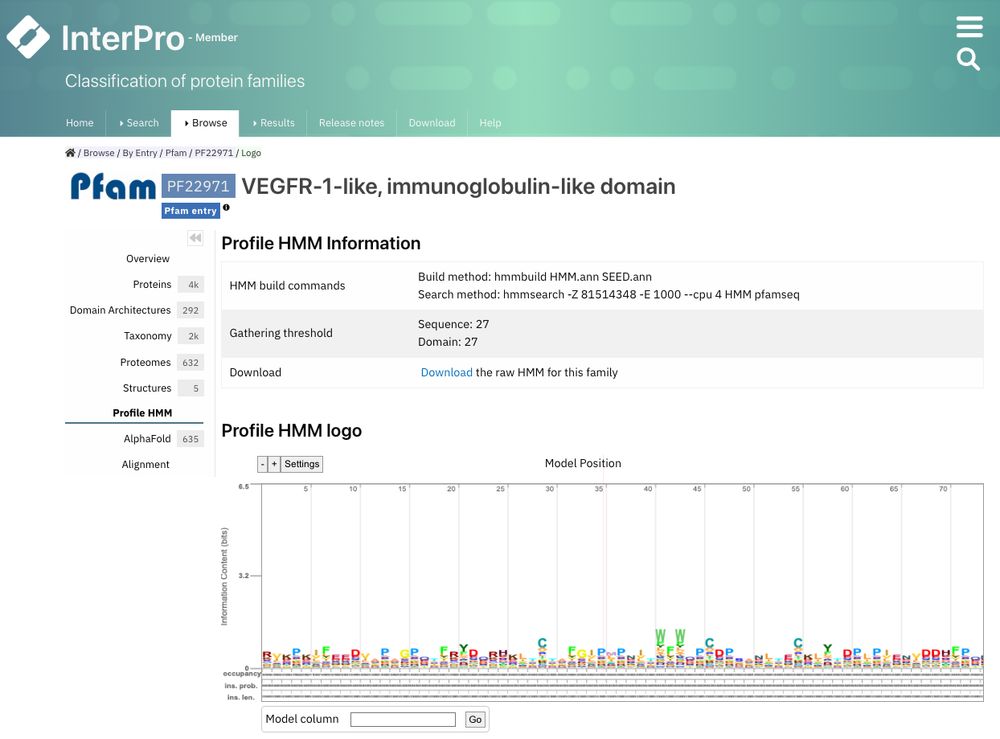
1) Curation data are now in Overview
2) HMM information and logo are now in Profile HMM
Have a look: www.ebi.ac.uk/interpro/ent...
These courses are currently open for applications - find out more: www.ebi.ac.uk/training/liv...
💻🧬💻🧪
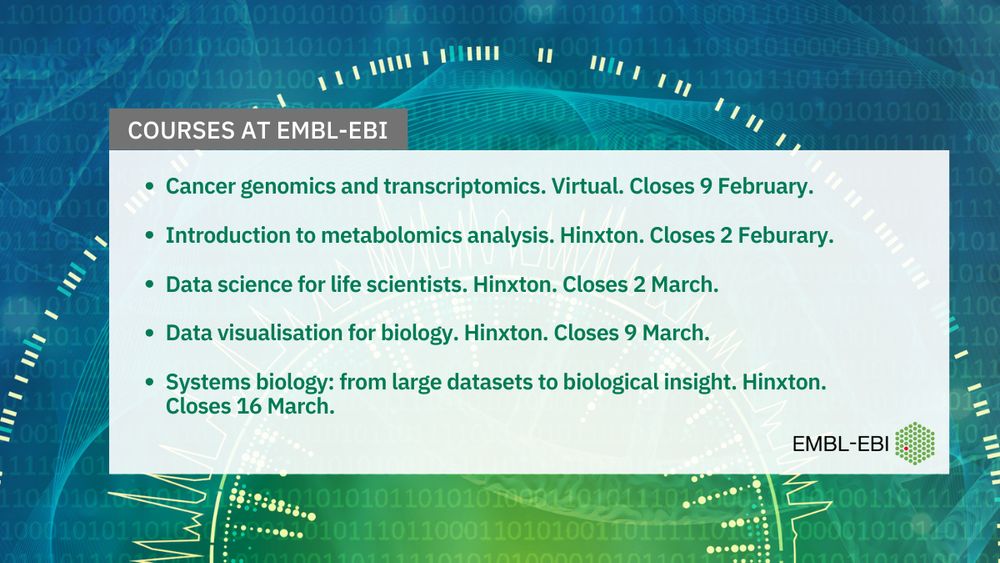
These courses are currently open for applications - find out more: www.ebi.ac.uk/training/liv...
💻🧬💻🧪
domains. doi.org/10.1093/bioa... 1/7
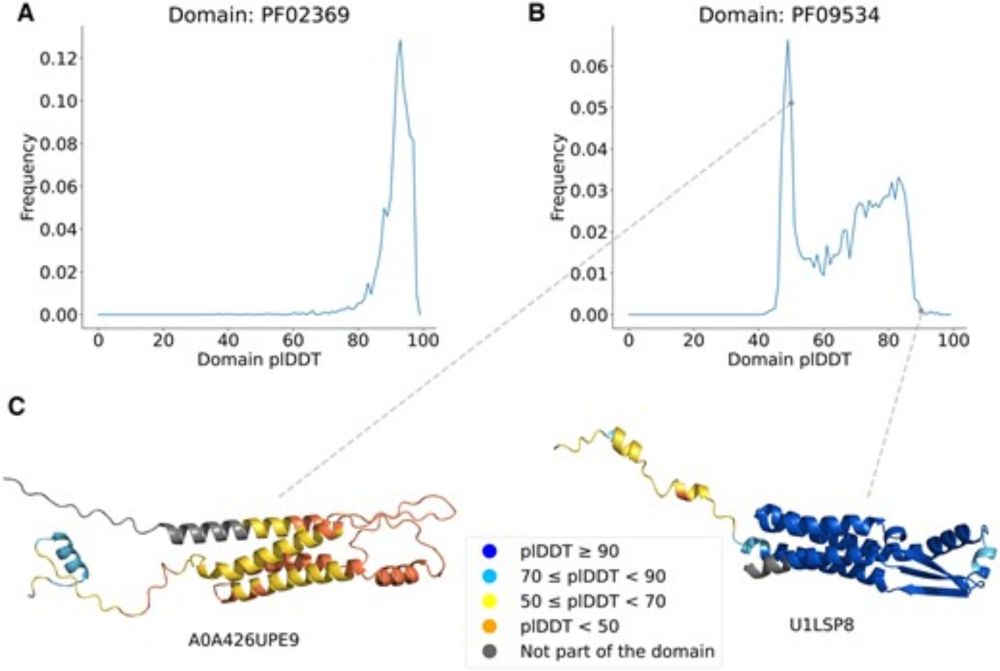
domains. doi.org/10.1093/bioa... 1/7
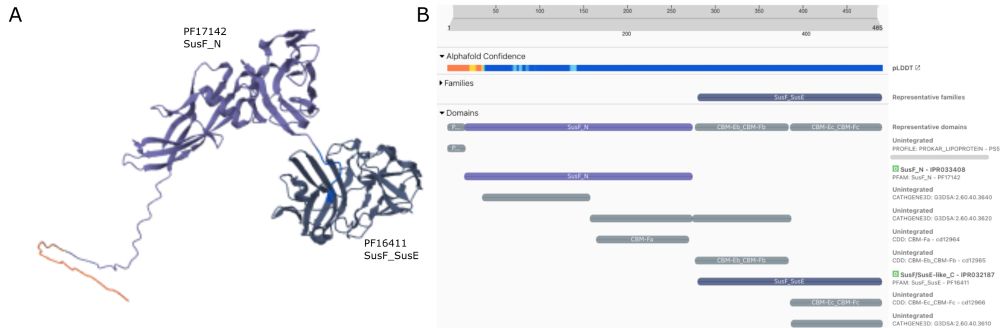
t.co/8auCxw3mdj
t.co/8auCxw3mdj
✨Update to Pfam 37.1
✨1029 new entries, including 1019 signatures from the Pfam (1008), NCBIfam (2), PIRSF (1), PANTHER (1), CDD (7) databases.
Have a look: ebi.ac.uk/interpro/




✨Update to Pfam 37.1
✨1029 new entries, including 1019 signatures from the Pfam (1008), NCBIfam (2), PIRSF (1), PANTHER (1), CDD (7) databases.
Have a look: ebi.ac.uk/interpro/


