- Founding partner of the wwPDB.
- Manage the community-led PDBe-KB resource and the AlphaFold Protein Structure Database, a collaboration with Google DeepMind.
- Part of EMBL-EBI
The PDBe wishes you a very Merry Christmas and a Happy New Year. We are delighted to share our jointly designed EMDB/PDBe Christmas card with you. It has been a busy year, and we look forward to seeing you in the New Year.

The PDBe wishes you a very Merry Christmas and a Happy New Year. We are delighted to share our jointly designed EMDB/PDBe Christmas card with you. It has been a busy year, and we look forward to seeing you in the New Year.
This reflects advances in cryo-EM, high-throughput crystallography, and AI-driven modelling, supported by expert wwPDB biocuration and open data sharing.
Read more: wwpdb.org/news/news?ye...

This reflects advances in cryo-EM, high-throughput crystallography, and AI-driven modelling, supported by expert wwPDB biocuration and open data sharing.
Read more: wwpdb.org/news/news?ye...
The new PDBe-KB Ligand pages are live with a cleaner layout, intuitive tabs, and a brand-new onboarding tour to help you get started.
Explore here 👉 pdbe.org/chem/STI

The new PDBe-KB Ligand pages are live with a cleaner layout, intuitive tabs, and a brand-new onboarding tour to help you get started.
Explore here 👉 pdbe.org/chem/STI
The release aligns AFDB with UniProt 2025_03 and introduces a redesigned entry page with integrated annotations, domains, summaries, and a 3D viewer.
📄 Read more: academic.oup.com/nar/advance-...

The release aligns AFDB with UniProt 2025_03 and introduces a redesigned entry page with integrated annotations, domains, summaries, and a 3D viewer.
📄 Read more: academic.oup.com/nar/advance-...
Our latest PDBe entry page update adds smoother browsing, better 3D selections, and direct links to page sections.
Examples:
Macromolecule:
www.ebi.ac.uk/pdbe/entry/p...
Ligand:
www.ebi.ac.uk/pdbe/entry/p...
Complex:
www.ebi.ac.uk/pdbe/entry/p...

Our latest PDBe entry page update adds smoother browsing, better 3D selections, and direct links to page sections.
Examples:
Macromolecule:
www.ebi.ac.uk/pdbe/entry/p...
Ligand:
www.ebi.ac.uk/pdbe/entry/p...
Complex:
www.ebi.ac.uk/pdbe/entry/p...
Starting Jan 2026, data depositions will be redistributed:
• PDBj → Oceania + most of Asia (except China)
• PDBc → China
• RCSB PDB → Americas
• PDBe → Europe & Africa

Starting Jan 2026, data depositions will be redistributed:
• PDBj → Oceania + most of Asia (except China)
• PDBc → China
• RCSB PDB → Americas
• PDBe → Europe & Africa
🔗 Explore (SARS-CoV-2 spike, PDB 9CXE): ebi.ac.uk/pdbe/pdbe-kb...
📄 Read the preprint: ssrn.com/abstract=568...
#PDBeKB #MacromolecularComplexes #StructuralBiology

🔗 Explore (SARS-CoV-2 spike, PDB 9CXE): ebi.ac.uk/pdbe/pdbe-kb...
📄 Read the preprint: ssrn.com/abstract=568...
#PDBeKB #MacromolecularComplexes #StructuralBiology

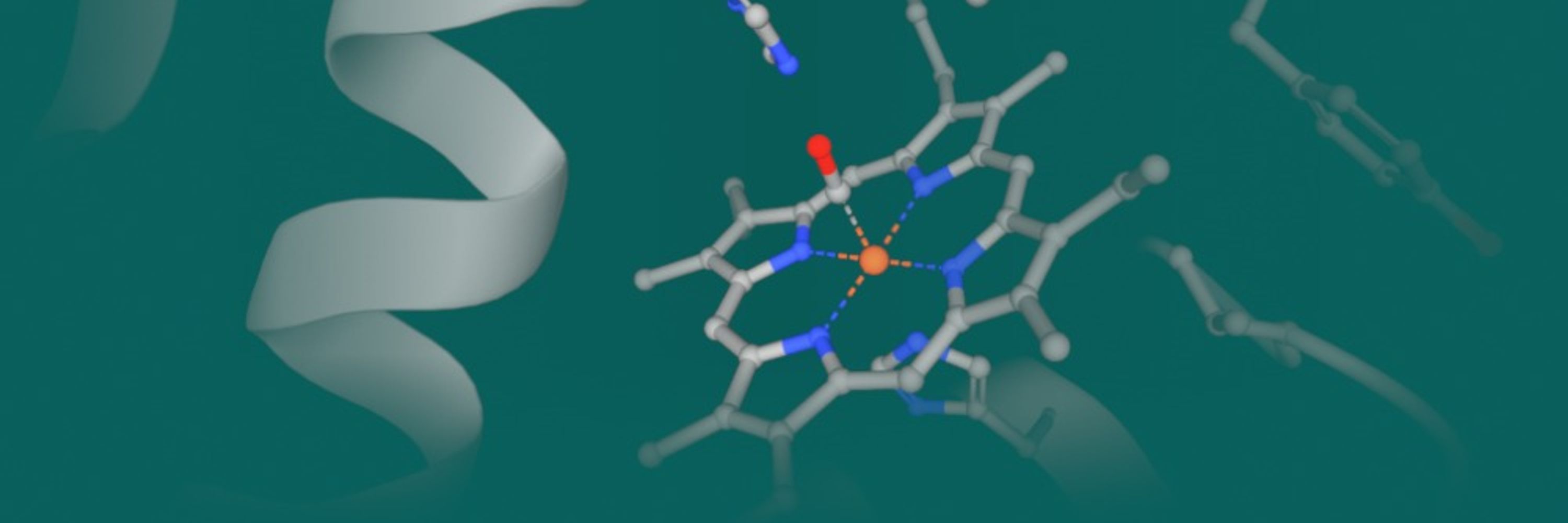
To ensure FAIR data practices, wwPDB is remediating existing polyatomic metal-containing entries and CCDs — improving their chemical and coordination descriptions for better findability and reuse.

To ensure FAIR data practices, wwPDB is remediating existing polyatomic metal-containing entries and CCDs — improving their chemical and coordination descriptions for better findability and reuse.
Old: alphafold.com/entry/P40763
New: alphafold.com/entry/AF-P40...
Simply add 'AF-' and '-F1' to the UniProt ID. Update your bookmarks and code!
Old: alphafold.com/entry/P40763
New: alphafold.com/entry/AF-P40...
Simply add 'AF-' and '-F1' to the UniProt ID. Update your bookmarks and code!
Totals v4 → v6: 241,070,489 entries.
Since v4: +65,711,653 added, 39,324,993 removed, 276,539 changed (in sequence), 175,082,297 unchanged (metadata refresh, relabelled v6). Details 👇

Totals v4 → v6: 241,070,489 entries.
Since v4: +65,711,653 added, 39,324,993 removed, 276,539 changed (in sequence), 175,082,297 unchanged (metadata refresh, relabelled v6). Details 👇
Find out more, register, submit your abstract ➡️ mddbr.eu/conference/
#MDDB #OpenScience #FAIRdata
📅 February 3-5, 2026
📍 Lausanne, Switzerland
Join us for an inspiring event with outstanding speakers and experts in the field!
Read more and register here ➡️ mddbr.eu/conference/
#MDDB #OpenScience #FAIRdata

Find out more, register, submit your abstract ➡️ mddbr.eu/conference/
#MDDB #OpenScience #FAIRdata
We'll keep developing the AlphaFold Database to support protein science worldwide 🎉
To mark the moment we’ve synchronised the database with UniProtKB release 2025_03.
www.ebi.ac.uk/about/news/t...
🖥️🧬 #AlphaFold
@pdbeurope.bsky.social

We'll keep developing the AlphaFold Database to support protein science worldwide 🎉
To mark the moment we’ve synchronised the database with UniProtKB release 2025_03.
www.ebi.ac.uk/about/news/t...
🖥️🧬 #AlphaFold
@pdbeurope.bsky.social
For more information, read the news item: www.ebi.ac.uk/pdbe/news/br...
For more information, read the news item: www.ebi.ac.uk/pdbe/news/br...
From Oct 1, all new 3DEM entries will include the Q-score percentile slider — helping users quickly assess model–map fit quality across EMDB/PDB archives.
Read more: www.wwpdb.org/news/news?ye...

From Oct 1, all new 3DEM entries will include the Q-score percentile slider — helping users quickly assess model–map fit quality across EMDB/PDB archives.
Read more: www.wwpdb.org/news/news?ye...
The new PDBe entry pages make it easier to explore protein structures.
🟢 Visualise ligands & macromolecules with improved 3D viewers
🟢 Get background info from AI-based annotations
🟢 Add your own annotations to viewers without leaving the browser
@pdbeurope.bsky.social
🧬🖥️
The new PDBe entry pages make it easier to explore protein structures.
🟢 Visualise ligands & macromolecules with improved 3D viewers
🟢 Get background info from AI-based annotations
🟢 Add your own annotations to viewers without leaving the browser
@pdbeurope.bsky.social
🧬🖥️
✨ Discover the new interactive, personalised & mobile-ready PDBe Entry Pages.
📖 Read more: www.ebi.ac.uk/about/news/u...
🔍 Try it: www.ebi.ac.uk/pdbe/entry/p...

✨ Discover the new interactive, personalised & mobile-ready PDBe Entry Pages.
📖 Read more: www.ebi.ac.uk/about/news/u...
🔍 Try it: www.ebi.ac.uk/pdbe/entry/p...
Try the new PDBe-KB Complex pages and let us know:
🔸 What’s working?
🔸 What’s confusing?
🔸 What features are lacking
💬 Feedback form: tinyurl.com/pdbekb-compl...
#PDBeKB #Complexes #Bioinformatics #StructuralBiology #Science

Try the new PDBe-KB Complex pages and let us know:
🔸 What’s working?
🔸 What’s confusing?
🔸 What features are lacking
💬 Feedback form: tinyurl.com/pdbekb-compl...
#PDBeKB #Complexes #Bioinformatics #StructuralBiology #Science
🗂️ Unified view of PDB assemblies
🧬 Standardised annotations
📊 Structural metrics and interactive visualisations
🔗 wwwdev.ebi.ac.uk/pdbe/pdbe-kb...
#PDBeKB #Complexes #Bioinformatics #StructuralBiology #Science

🗂️ Unified view of PDB assemblies
🧬 Standardised annotations
📊 Structural metrics and interactive visualisations
🔗 wwwdev.ebi.ac.uk/pdbe/pdbe-kb...
#PDBeKB #Complexes #Bioinformatics #StructuralBiology #Science
Fill in a 6-minute survey about your experience with PDBePISA
forms.gle/PfoCYHJe1y9L...

Fill in a 6-minute survey about your experience with PDBePISA
forms.gle/PfoCYHJe1y9L...
Same macromolecular complex. Different experimental conditions. All assemblies in one place.
🔍 Try the beta now: wwwdev.ebi.ac.uk/pdbe/pdbe-kb...
#PDBeKB #Complexes #Bioinformatics #StructuralBiology #Science

Same macromolecular complex. Different experimental conditions. All assemblies in one place.
🔍 Try the beta now: wwwdev.ebi.ac.uk/pdbe/pdbe-kb...
#PDBeKB #Complexes #Bioinformatics #StructuralBiology #Science
PDBe brings new updated REST API endpoints that fully replicates the functionality of the old service, while introducing performance enhancements and additional endpoints.
⬇️ Read more⬇️
www.ebi.ac.uk/pdbe/news/un...

PDBe brings new updated REST API endpoints that fully replicates the functionality of the old service, while introducing performance enhancements and additional endpoints.
⬇️ Read more⬇️
www.ebi.ac.uk/pdbe/news/un...
🧪 MS6_P7 - Building a FAIR Repository for Fragment-Based Screening Campaigns - Presented by Genevieve Evans
All ECM poster titles & abstracts available here:
🔗 ecm35.ecanews.org/poster-hiper...
#Biology #Chemistry
🧪 MS6_P7 - Building a FAIR Repository for Fragment-Based Screening Campaigns - Presented by Genevieve Evans
All ECM poster titles & abstracts available here:
🔗 ecm35.ecanews.org/poster-hiper...
#Biology #Chemistry
2026 PDBArt calendars 🎨🗓️ available.
🔗 www.ebi.ac.uk/pdbe/pdb-art
Crystallography continues to unlock the secrets of biomolecular structures!
#Biology # Chemistry
2026 PDBArt calendars 🎨🗓️ available.
🔗 www.ebi.ac.uk/pdbe/pdb-art
Crystallography continues to unlock the secrets of biomolecular structures!
#Biology # Chemistry
We are organising a 1-day symposium on September 16th at UCL, highlighting recent AI-based developments to enhance protein family classifications, annotations and analyses.
www.eventbrite.co.uk/e/protein-an...

We are organising a 1-day symposium on September 16th at UCL, highlighting recent AI-based developments to enhance protein family classifications, annotations and analyses.
www.eventbrite.co.uk/e/protein-an...

