bit.ly/4od3ux8
Plenty of future SynBio/EngBio applications planned! Thanks to all involved (character limits suck).
bit.ly/4od3ux8
Plenty of future SynBio/EngBio applications planned! Thanks to all involved (character limits suck).
We mapped intramolecular isopeptide bonds across the AlphaFold database and found that they are widely distributed in microbial surface proteins, such as fibrillar adhesins or pilins, suggesting new targets for broad-spectrum antimicrobial strategies.
www.biorxiv.org/content/10.1...

We mapped intramolecular isopeptide bonds across the AlphaFold database and found that they are widely distributed in microbial surface proteins, such as fibrillar adhesins or pilins, suggesting new targets for broad-spectrum antimicrobial strategies.
academic.oup.com/bioinformati...

academic.oup.com/bioinformati...
Francesco is fascinated by protein design. His recent work helped ‘rescue’ low-confidence #AlphaFold protein structure predictions.
www.embl.org/news/people-...
🧬🧶#MolBiol
@ebi.embl.org

But things get more challenging for protein families.
A new method helps improve low confidence predictions within protein families.
academic.oup.com/bioinformati...
But things get more challenging for protein families.
A new method helps improve low confidence predictions within protein families.
academic.oup.com/bioinformati...
domains. doi.org/10.1093/bioa... 1/7
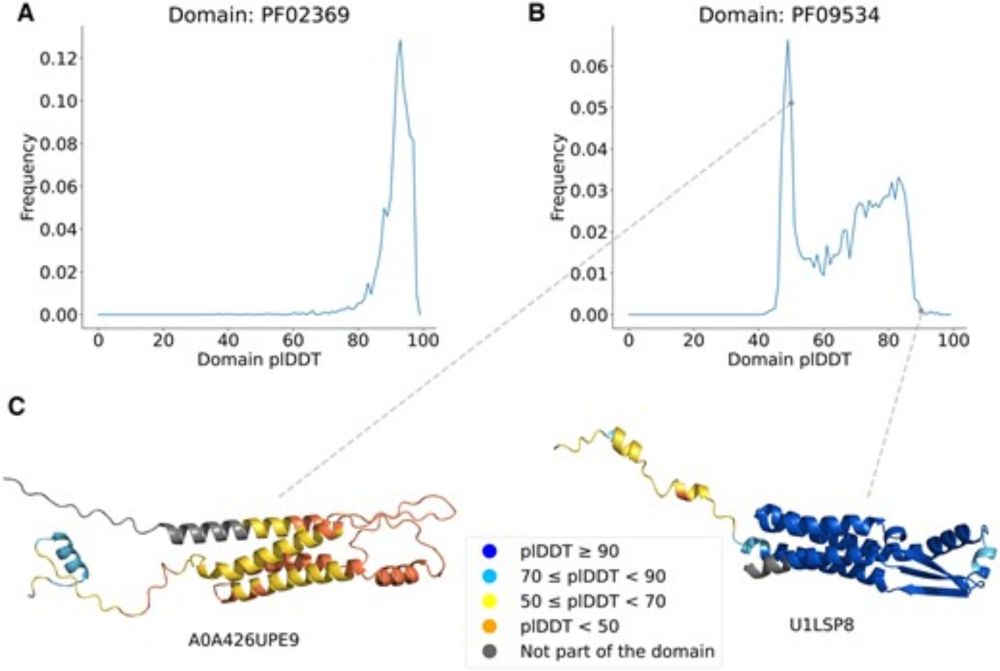
domains. doi.org/10.1093/bioa... 1/7

