CNRS, University of Montpellier, France.
https://www.igh.cnrs.fr/en/project/index/0/134/igh
Enjoy!!!
repairome.bioinfo.cnio.es
Enjoy!!!
repairome.bioinfo.cnio.es
genomebiology.biomedcentral.com/articles/10....
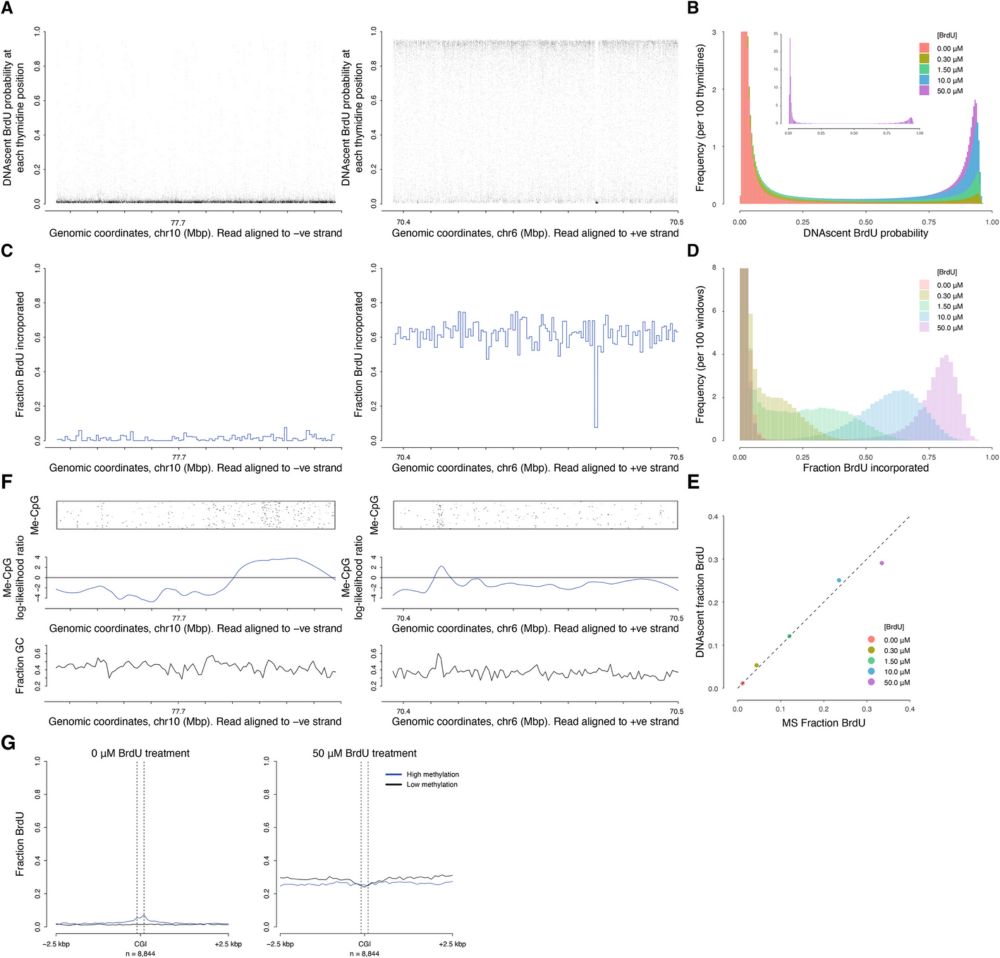
Identifying human DNA replication initiation sites on single molecules by @nanoporetech.com 1/7
genomebiology.biomedcentral.com/articles/10....
Identifying human DNA replication initiation sites on single molecules by @nanoporetech.com 1/7
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
ATM controls fork processing and restart, and the PPM1D phosphatase is needed to properly balance this action of ATM.
Congratulations to @yitingcao.bsky.social @yingzhengwang.bsky.social and Jumana Badar.
www.biorxiv.org/content/10.1...

ATM controls fork processing and restart, and the PPM1D phosphatase is needed to properly balance this action of ATM.
Congratulations to @yitingcao.bsky.social @yingzhengwang.bsky.social and Jumana Badar.
www.biorxiv.org/content/10.1...
Spoiler: scientists love it.
Huge thanks to the amazing team at The Company of Biologists for making Fast & Fair happen.
Read the preprint: bit.ly/4iZ30st
#biologists100 @biologists.bsky.social @biologyopen.bsky.social

Spoiler: scientists love it.
Huge thanks to the amazing team at The Company of Biologists for making Fast & Fair happen.
Read the preprint: bit.ly/4iZ30st
#biologists100 @biologists.bsky.social @biologyopen.bsky.social
Contact: nestor.garcia@cabimer.es

Contact: nestor.garcia@cabimer.es



@jama.com
jamanetwork.com/journals/jam...
@plos.org
theplosblog.plos.org/2025/02/plos...
@science.org
www.science.org/doi/10.1126/...
@nature.com
www.nature.com/articles/d41...
@embopress.org
www.embopress.org/doi/full/10....
@jama.com
jamanetwork.com/journals/jam...
@plos.org
theplosblog.plos.org/2025/02/plos...
@science.org
www.science.org/doi/10.1126/...
@nature.com
www.nature.com/articles/d41...
@embopress.org
www.embopress.org/doi/full/10....




Learn more here:
➡️ tinyurl.com/gd352227
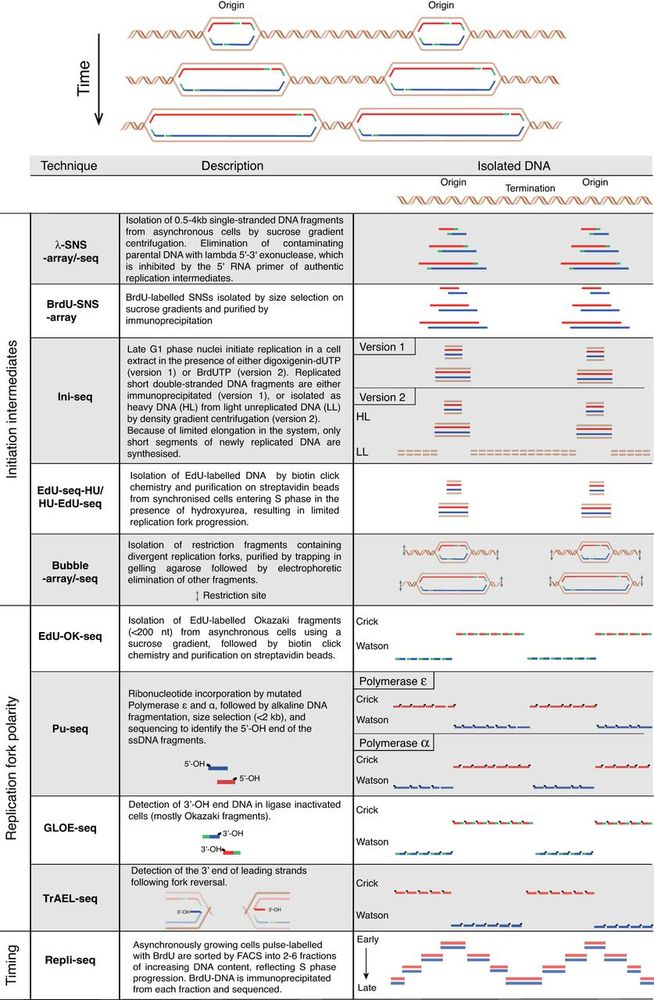
Learn more here:
➡️ tinyurl.com/gd352227
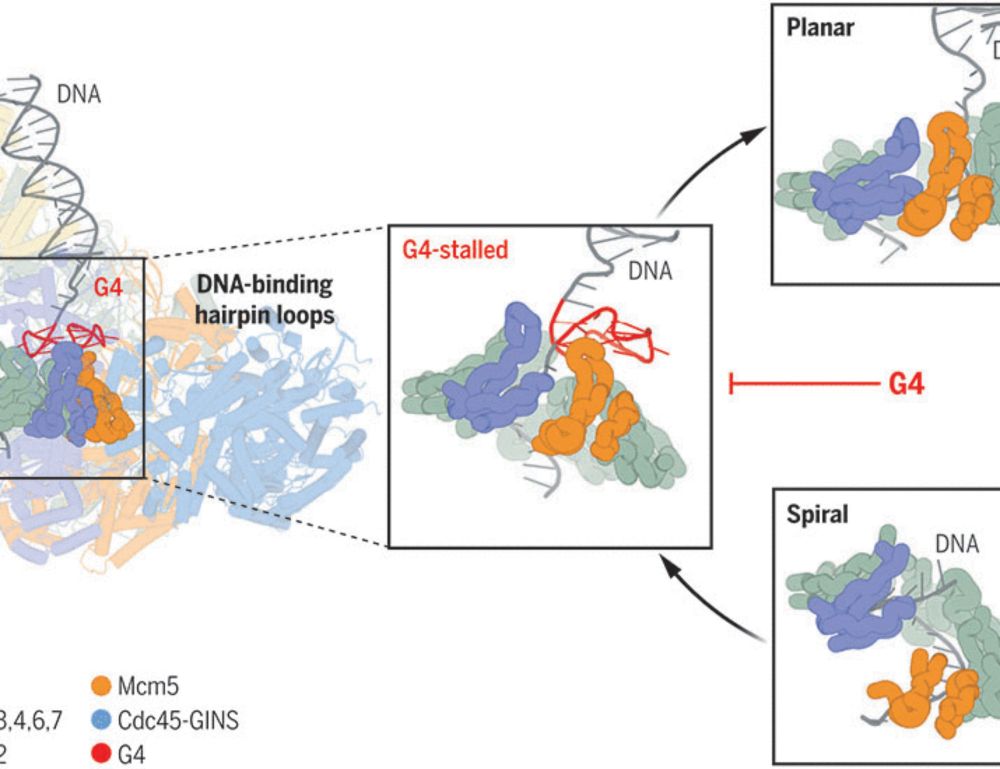

Organized by Helle Ulrich (IMB), Johannes Walter (Harvard) and Anja Groth (Danish Cancer Institute).
www.embl.org/about/info/c...
Organized by Helle Ulrich (IMB), Johannes Walter (Harvard) and Anja Groth (Danish Cancer Institute).
www.embl.org/about/info/c...
📆May 28-30th
👥 @oscarllorca.bsky.social, @rfleiro.bsky.social and @nogaleslab.bsky.social.
🔗https://www.cnio.es/eventos/machines-acting-on-dna-and-rna-a-molecular-mechanistic-perspective/

📆May 28-30th
👥 @oscarllorca.bsky.social, @rfleiro.bsky.social and @nogaleslab.bsky.social.
🔗https://www.cnio.es/eventos/machines-acting-on-dna-and-rna-a-molecular-mechanistic-perspective/
@MorieLAB.bsky.social!
Neutral Lipids restrict DNA mobility!!
1. This has important implications for DNA repair in vivo,
2. also in vitro when you measure DNA breaks using the comet assay, beware!
3. And, we provide a fast new protocol overcoming this issue 😍
doi.org/10.1111/boc....

@MorieLAB.bsky.social!
Neutral Lipids restrict DNA mobility!!
1. This has important implications for DNA repair in vivo,
2. also in vitro when you measure DNA breaks using the comet assay, beware!
3. And, we provide a fast new protocol overcoming this issue 😍
doi.org/10.1111/boc....
Kara Bernstein, Katrin Paeschke and coworkers
www.embopress.org/doi/full/10....

Kara Bernstein, Katrin Paeschke and coworkers
www.embopress.org/doi/full/10....
Mec1 (yeast ATR) can’t target D/E-S/T, so we think D/E-S/T targeting allows Tel1 functional specialization.
A mutation in Tel1 makes it only target S/T-Q… and cells become CPT sensitive.

Mec1 (yeast ATR) can’t target D/E-S/T, so we think D/E-S/T targeting allows Tel1 functional specialization.
A mutation in Tel1 makes it only target S/T-Q… and cells become CPT sensitive.

Nice work by Ismail Hassan Ismail lab in Canada!
www.cell.com/cell-reports...

Nice work by Ismail Hassan Ismail lab in Canada!
www.cell.com/cell-reports...

