https://pablormier.github.io/
->Nature | #Data | More info from EcoSearch

We present an extended version of ScAPE, the method that won one of the prizes 🏆 in the @neuripsconf.bsky.social 2023 Single-Cell Perturbation Prediction challenge.
📄 preprint: doi.org/10.1101/2025...
🧬 code: github.com/scapeML/scape

We present an extended version of ScAPE, the method that won one of the prizes 🏆 in the @neuripsconf.bsky.social 2023 Single-Cell Perturbation Prediction challenge.
📄 preprint: doi.org/10.1101/2025...
🧬 code: github.com/scapeML/scape
🔗
www.biorxiv.org/content/10.1...
📦
saezlab.github.io/MetaProViz/
🧵 Thread ⬇️

🔗
www.biorxiv.org/content/10.1...
📦
saezlab.github.io/MetaProViz/
🧵 Thread ⬇️
We're thrilled to be hosting the @embo.org Practical Course 'Causality in biomedicine: going beyond associations' from 4 – 9 October 2026.
Register your interest and be the first to hear when the course opens for applications: www.ebi.ac.uk/training/eve...

We're thrilled to be hosting the @embo.org Practical Course 'Causality in biomedicine: going beyond associations' from 4 – 9 October 2026.
Register your interest and be the first to hear when the course opens for applications: www.ebi.ac.uk/training/eve...
Very much looking forward to teaching and discussing!
We're thrilled to be hosting the @embo.org Practical Course 'Causality in biomedicine: going beyond associations' from 4 – 9 October 2026.
Register your interest and be the first to hear when the course opens for applications: www.ebi.ac.uk/training/eve...

Very much looking forward to teaching and discussing!
🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇
🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇

🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇
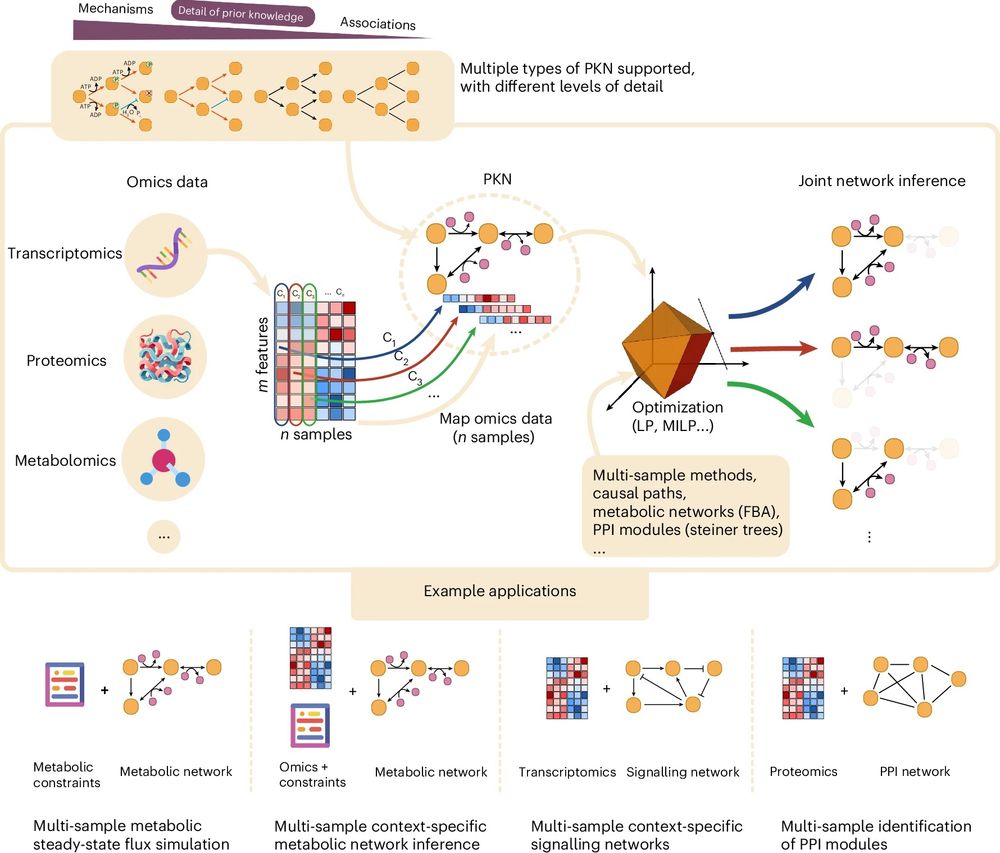
CORNETO is an open-source tool that uses machine learning to turn tangled omics datasets into clear maps of how genes, proteins, and signalling pathways interact.
www.ebi.ac.uk/about/news/r... 🧪
CORNETO is an open-source tool that uses machine learning to turn tangled omics datasets into clear maps of how genes, proteins, and signalling pathways interact.
www.ebi.ac.uk/about/news/r... 🧪
We present our new alignment framework TOAST www.biorxiv.org/content/10.1...
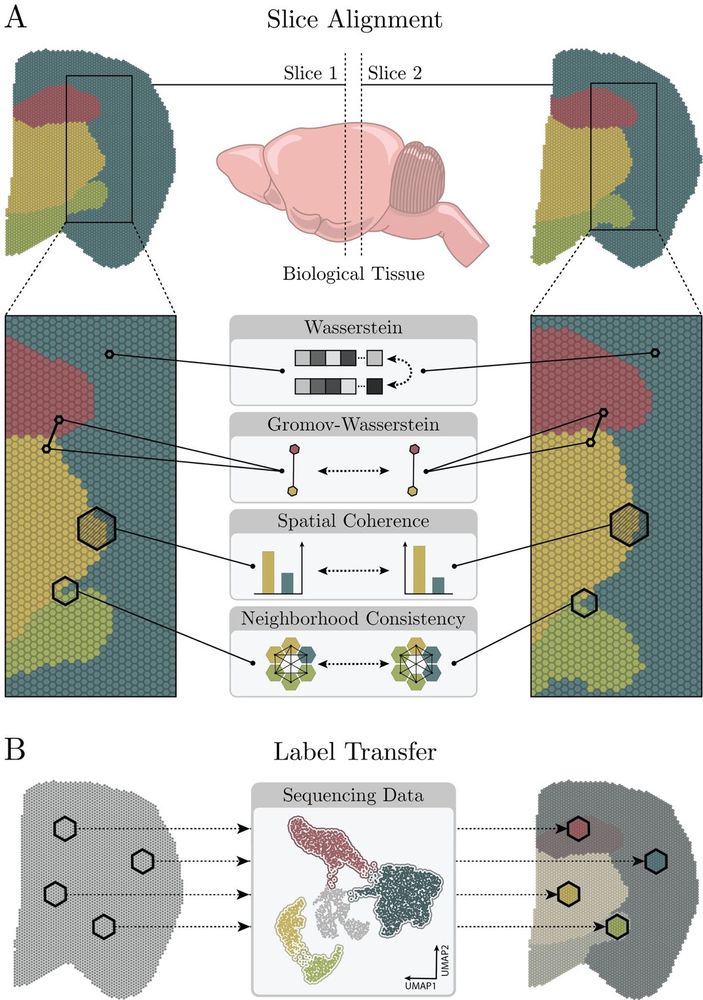
We present our new alignment framework TOAST www.biorxiv.org/content/10.1...
#virtualcells #foundationalmodels #compbio
blog.turbine.ai/p/pretrainin...

#virtualcells #foundationalmodels #compbio
blog.turbine.ai/p/pretrainin...
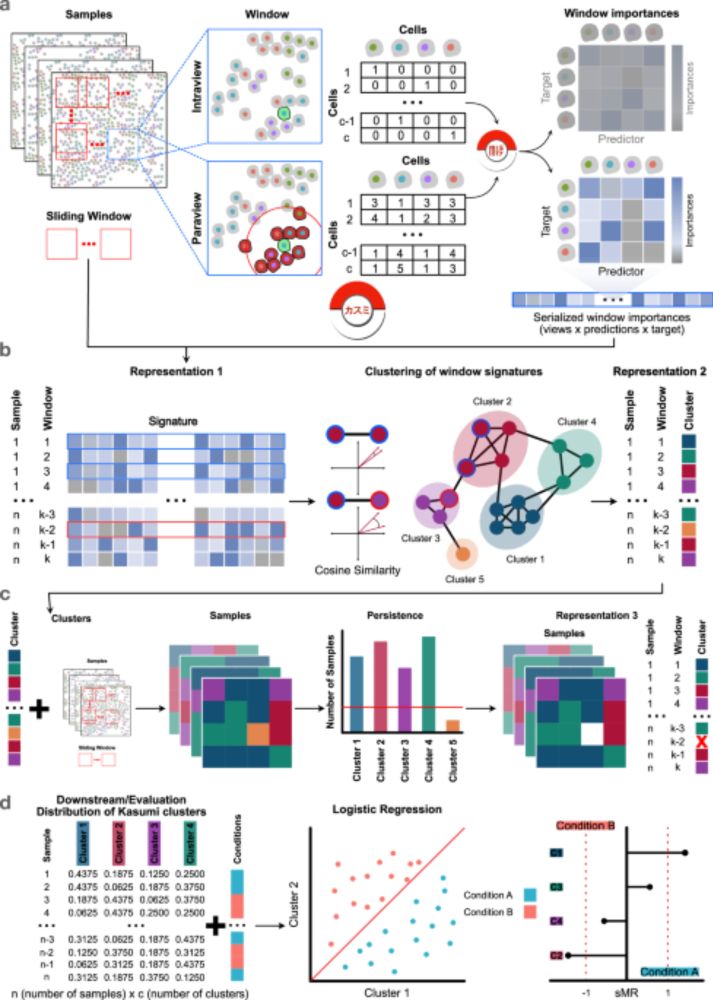
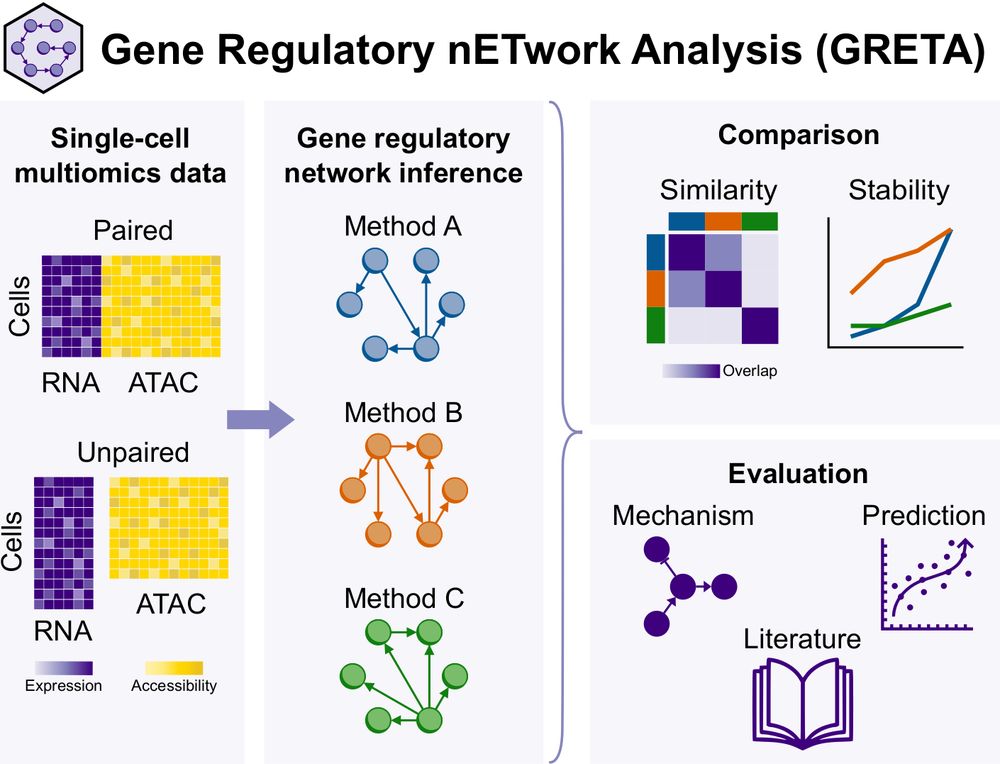
We have included the original and decoupled version of SCENIC+, added a new metric and two more databases. Dictys and SCENIC+ outperformed others, but still performed poorly in causal mechanistic tasks.
doi.org/10.1101/2024... 👇

We have included the original and decoupled version of SCENIC+, added a new metric and two more databases. Dictys and SCENIC+ outperformed others, but still performed poorly in causal mechanistic tasks.
doi.org/10.1101/2024... 👇
Fastest block ever!


Fastest block ever!
youtu.be/kV1ru-Inzl4?...

youtu.be/kV1ru-Inzl4?...
We need a LLM LaTeX benchmark!
We need a LLM LaTeX benchmark!
Register to participate: benchmarks.elsa-ai.eu?ch=4
Register to participate: benchmarks.elsa-ai.eu?ch=4
Paper: doi.org/10.1101/2024.12.20.629764
Code: github.com/saezlab/greta
Paper: doi.org/10.1101/2024.12.20.629764
Code: github.com/saezlab/greta
Paper: doi.org/10.1101/2024.12.20.629764
Code: github.com/saezlab/greta

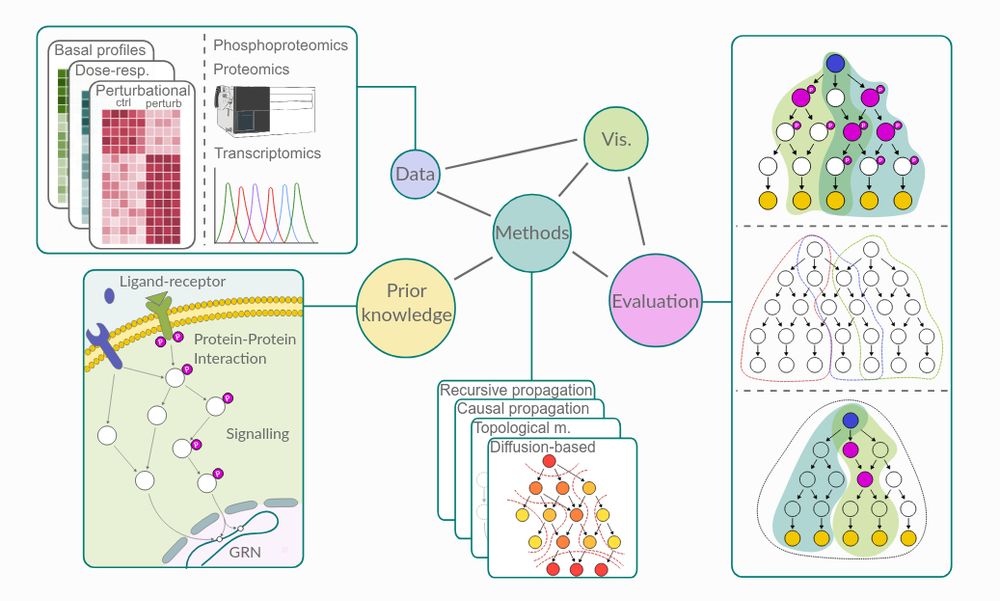
Paper: doi.org/10.1101/2024...
Docs: networkcommons.readthedocs.io
Paper: doi.org/10.1101/2024...
Docs: networkcommons.readthedocs.io
openreview.net/forum?id=WTI...
Thanks to the organizers for this amazing competition.

openreview.net/forum?id=WTI...
Thanks to the organizers for this amazing competition.


