
Preprint: www.medrxiv.org/content/10.1...

A team from the Cowley, Bassett, and @gosiatrynka.bsky.social labs integrate pooled iPSC phenotyping, transcriptomics, and CRISPR perturbations to identify genetic drivers of molecular and cellular phenotypes in microglia
www.biorxiv.org/content/10.1...
#neuroskyence

A team from the Cowley, Bassett, and @gosiatrynka.bsky.social labs integrate pooled iPSC phenotyping, transcriptomics, and CRISPR perturbations to identify genetic drivers of molecular and cellular phenotypes in microglia
www.biorxiv.org/content/10.1...
#neuroskyence
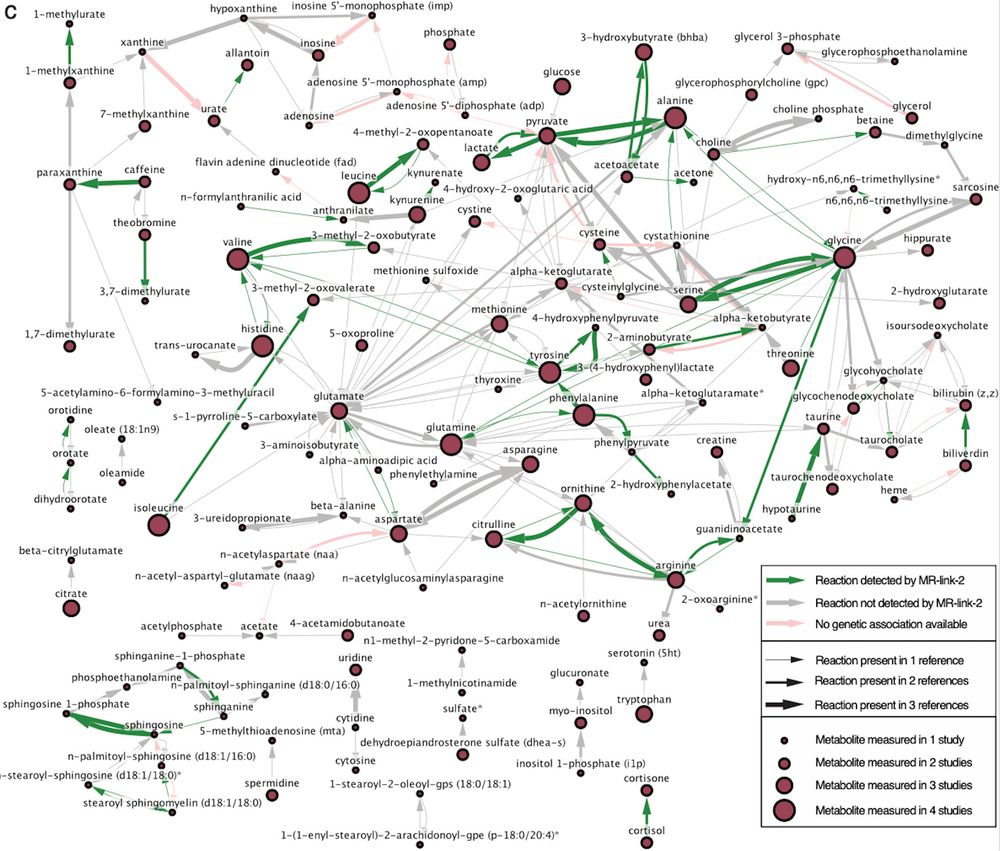
www.medrxiv.org/content/10.1...

www.medrxiv.org/content/10.1...
A team led by @tobioinformatics.bsky.social and Bradley Harris in @carlanderson.bsky.social ‘s lab has created the largest single-cell atlas of IBD tissues to date
www.medrxiv.org/content/10.1...
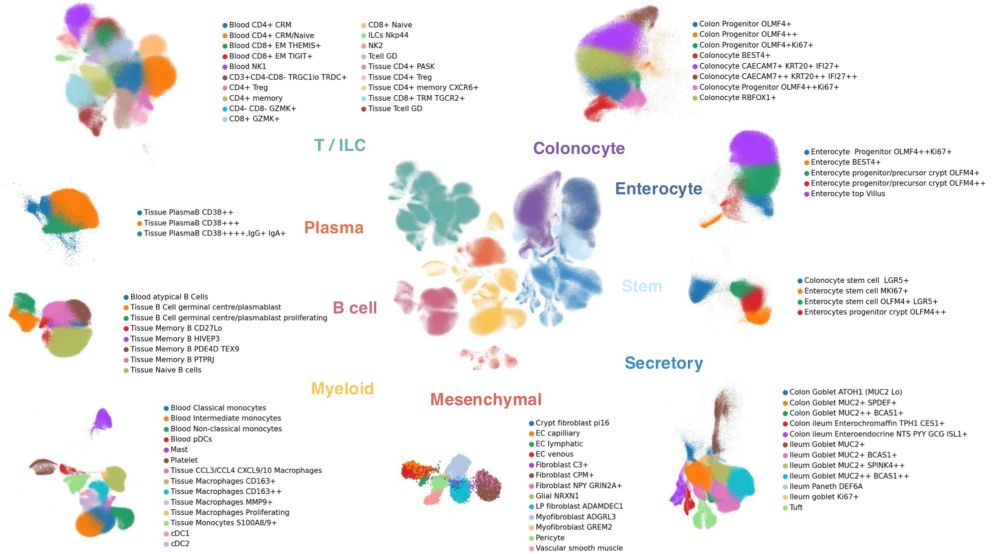
A team led by @tobioinformatics.bsky.social and Bradley Harris in @carlanderson.bsky.social ‘s lab has created the largest single-cell atlas of IBD tissues to date
www.medrxiv.org/content/10.1...
What do we mean when we say a regulatory effect is "context-specific" and how do we define the context?
We explored these questions using brain organoids and oxygen stress.
What do we mean when we say a regulatory effect is "context-specific" and how do we define the context?
We explored these questions using brain organoids and oxygen stress.
We expanded our study on parent-of-origin effects with new findings from the MoBa cohort, now leveraging up to 265,000 individuals!
Discover fresh insights into the genetic architecture of early growth!
Updated preprint: www.medrxiv.org/content/10.1...
With our novel method, we inferred the parental origin of alleles for >220,000 individuals, uncovering novel insights into genetics, evolution, and health.
👉 Read here: www.medrxiv.org/content/10.1...
👇 Highlights below!

We expanded our study on parent-of-origin effects with new findings from the MoBa cohort, now leveraging up to 265,000 individuals!
Discover fresh insights into the genetic architecture of early growth!
Updated preprint: www.medrxiv.org/content/10.1...
www.cell.com/cell-genomic...
Led by the talented @Charles_Zhou12 and supervised by @MengjieChen6
and me, with thanks to many contributors.
scPrediXcan integrates deep learning and single cell expression data into a powerful cell type specific TWAS framework.
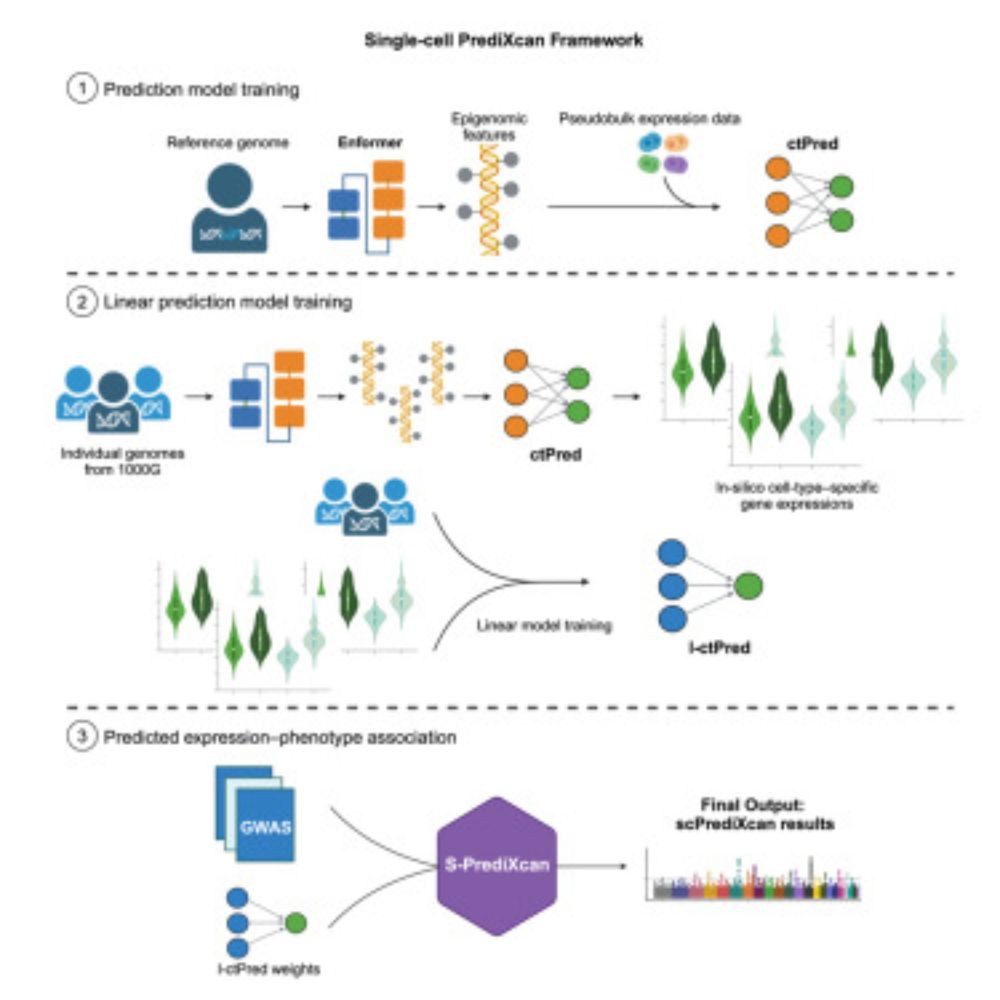
www.cell.com/cell-genomic...
Led by the talented @Charles_Zhou12 and supervised by @MengjieChen6
and me, with thanks to many contributors.
scPrediXcan integrates deep learning and single cell expression data into a powerful cell type specific TWAS framework.
Loads of cool findings including MR for alcohol usage -> increased variance in BC traits
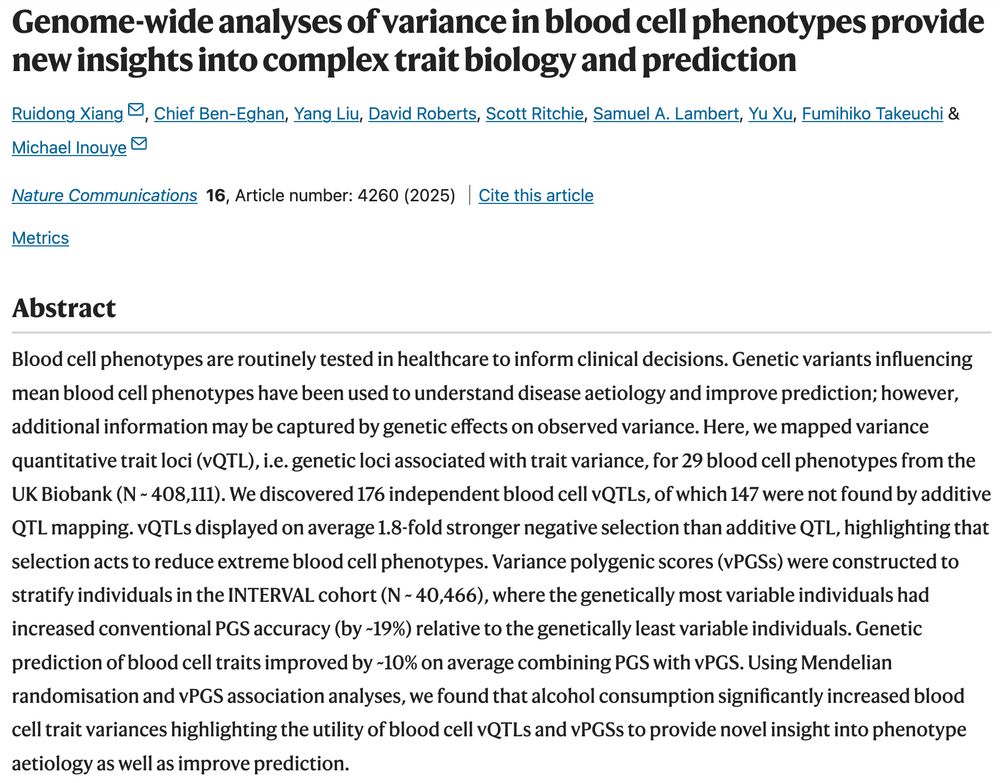
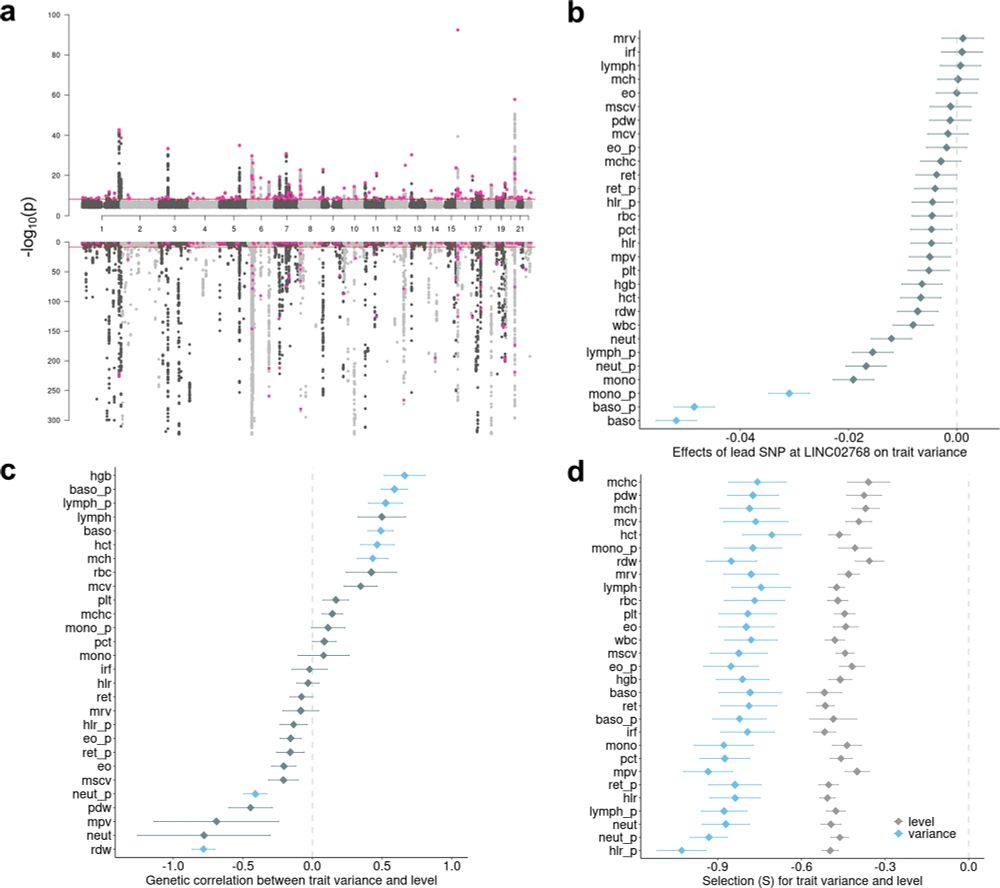
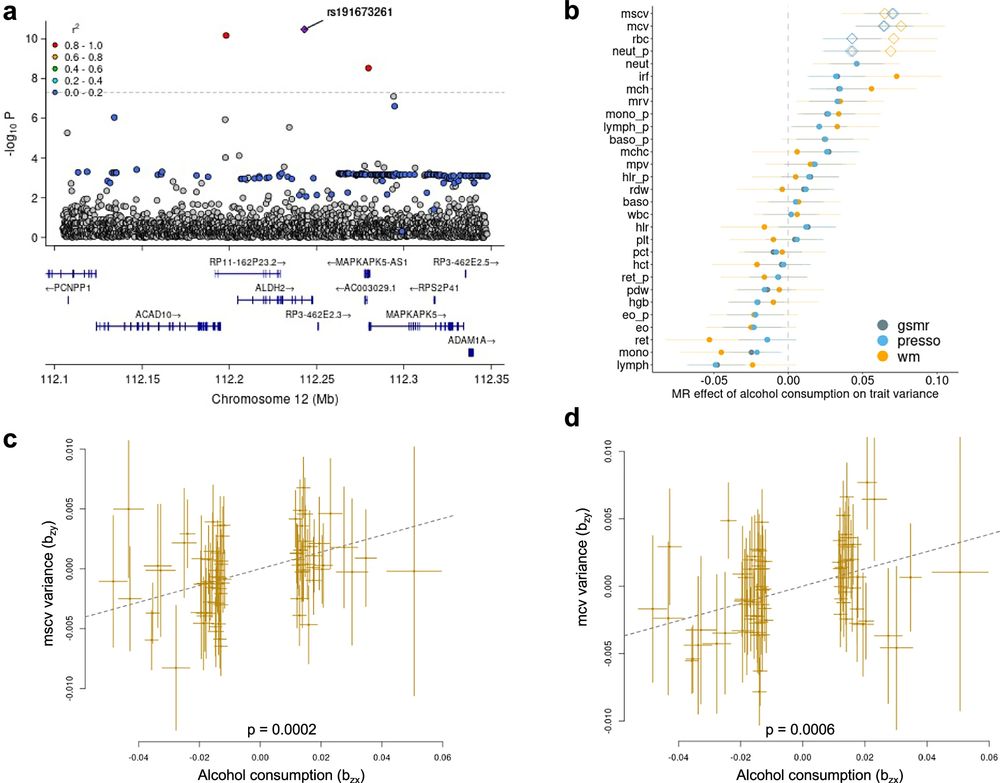
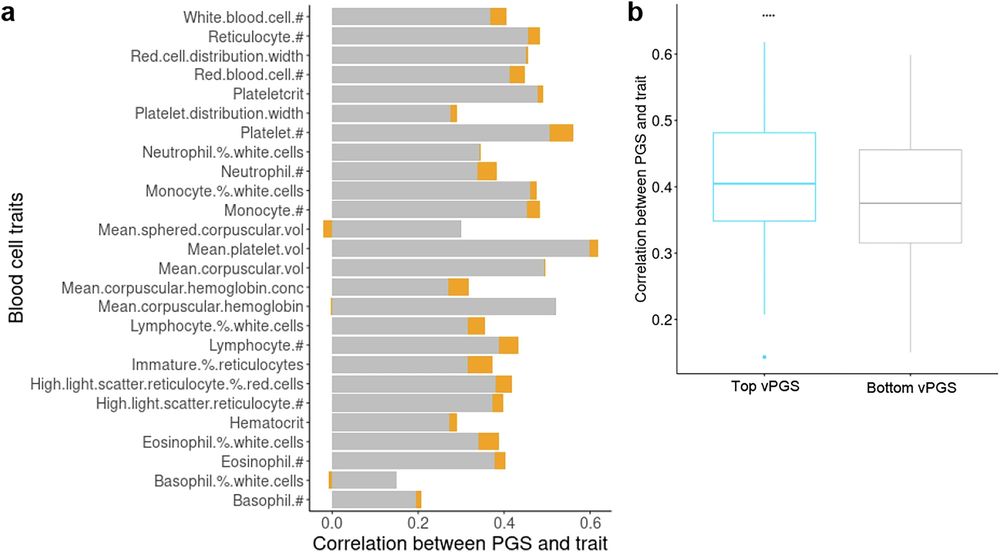
Loads of cool findings including MR for alcohol usage -> increased variance in BC traits

www.medrxiv.org/content/10.1... @medrxivpreprint.bsky.social




www.medrxiv.org/content/10.1... @medrxivpreprint.bsky.social
www.nature.com/articles/ng1...

www.nature.com/articles/ng1...
🧵👇 (1/n)
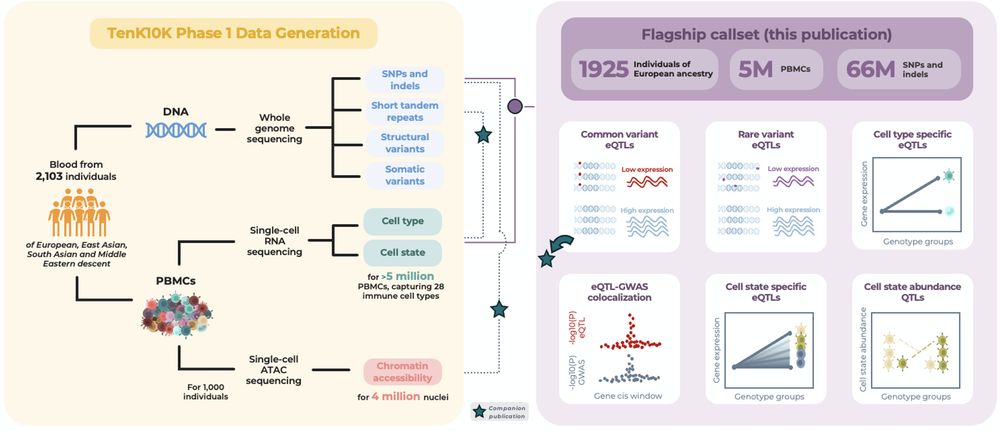
🧵👇 (1/n)
* Array genetic data for 80,638 Japanese children
* 1,163 child health and developmental traits (e.g. food allergy, anthropometric, developmental)
* Parental environmental exposures
www.medrxiv.org/content/10.1...

* Array genetic data for 80,638 Japanese children
* 1,163 child health and developmental traits (e.g. food allergy, anthropometric, developmental)
* Parental environmental exposures
www.medrxiv.org/content/10.1...
bsky.app/profile/nats...
Preprint: www.medrxiv.org/content/10.1...

bsky.app/profile/nats...
Check out the INTERVAL RNAseq portal www.intervalrna.org.uk
Led by @alextokolyi.bsky.social & Elodie Persyn!
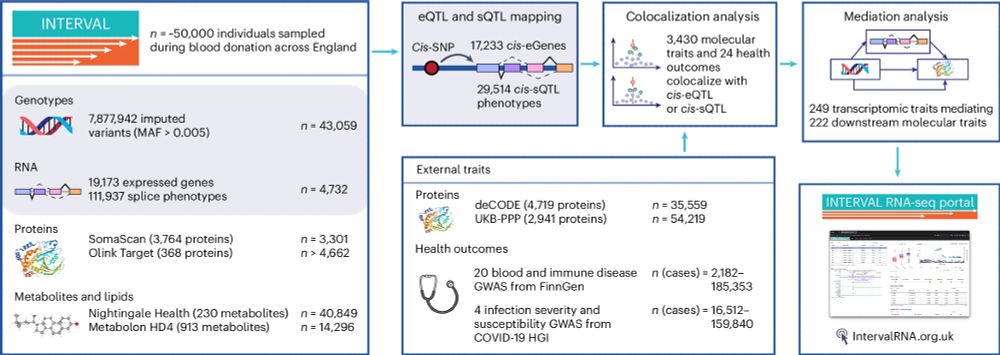
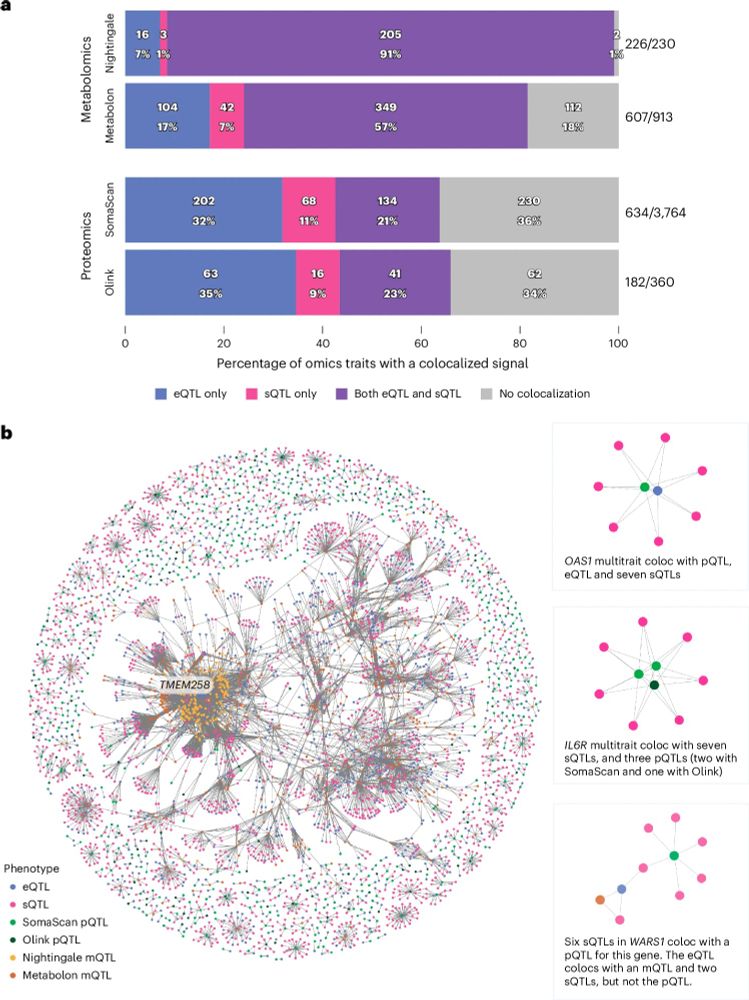
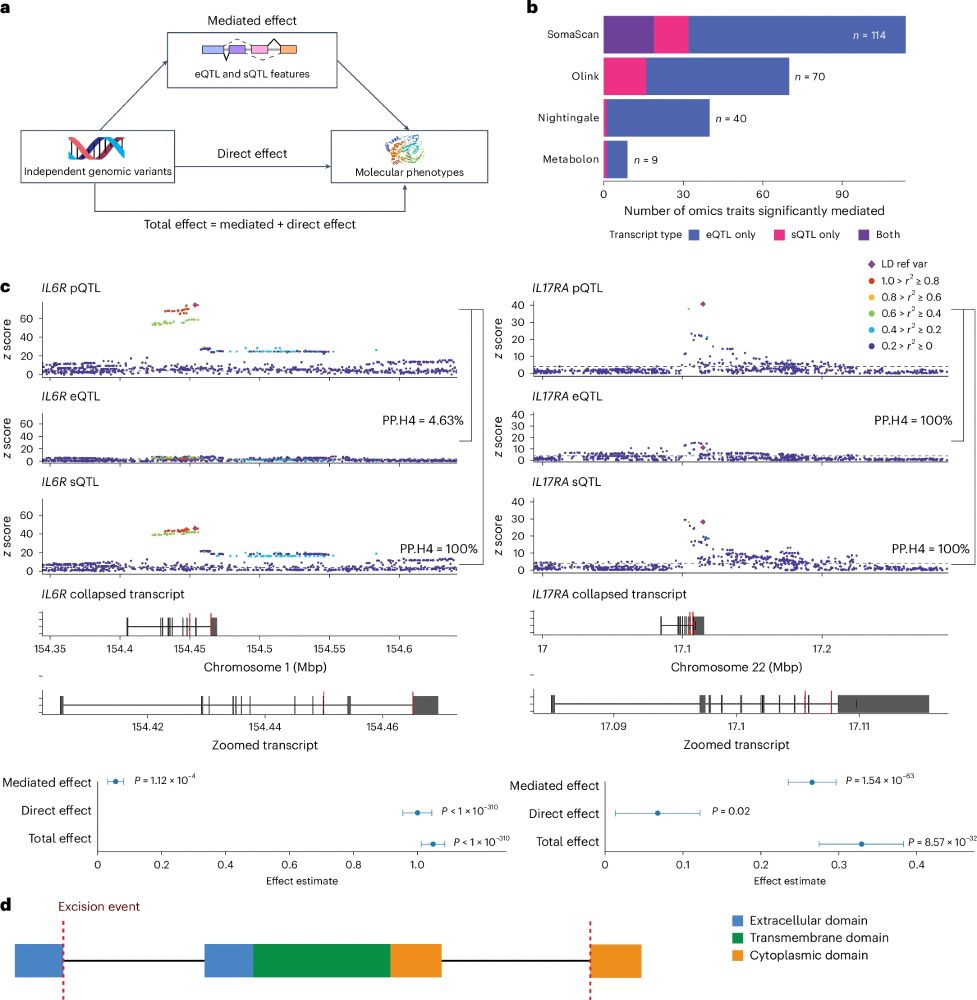
Check out the INTERVAL RNAseq portal www.intervalrna.org.uk
Led by @alextokolyi.bsky.social & Elodie Persyn!
From conventional machine learning methods to CNNs and using models as oracles/generative AI for synthetic enhancer design!
@natrevbioeng.bsky.social
www.nature.com/articles/s44...
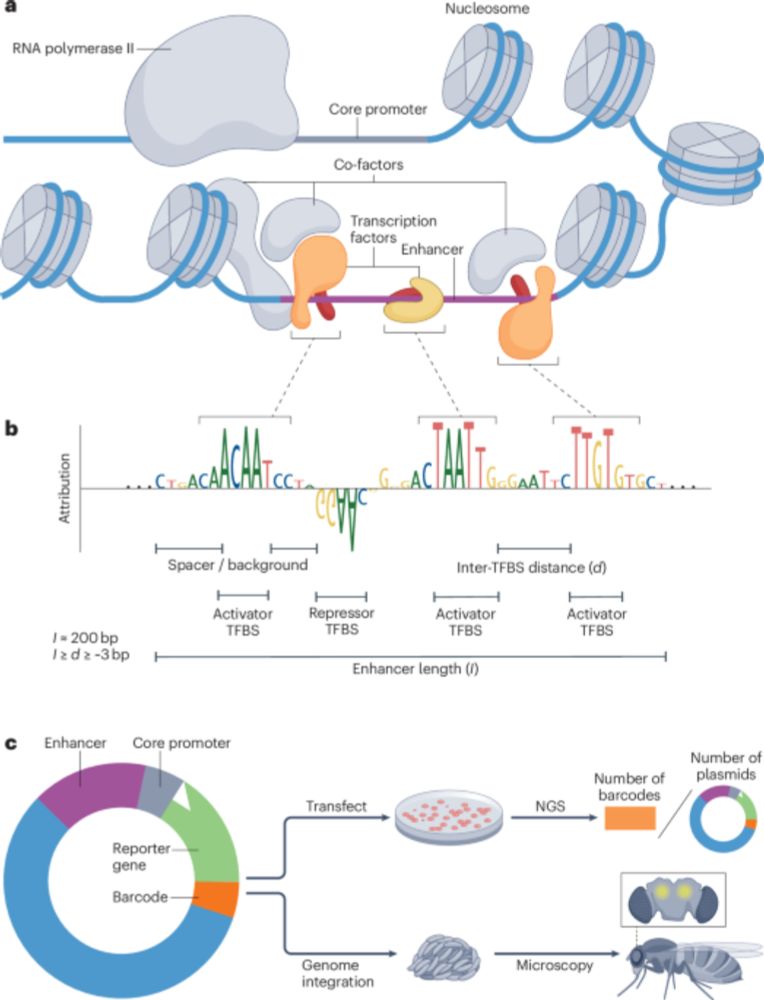
From conventional machine learning methods to CNNs and using models as oracles/generative AI for synthetic enhancer design!
@natrevbioeng.bsky.social
www.nature.com/articles/s44...

I'm very excited to present our new work combining associations and Perturb-seq to build interpretable causal graphs! A 🧵
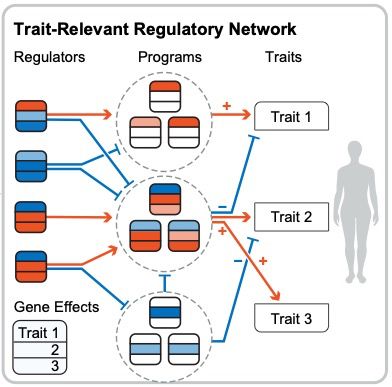
I'm very excited to present our new work combining associations and Perturb-seq to build interpretable causal graphs! A 🧵
Grateful to the #TOPMed #Omics WG for giving me an opportunity to learn and contribute to this important work 🧬

Grateful to the #TOPMed #Omics WG for giving me an opportunity to learn and contribute to this important work 🧬
Preprint: www.medrxiv.org/content/10.1...

Preprint: www.medrxiv.org/content/10.1...





