
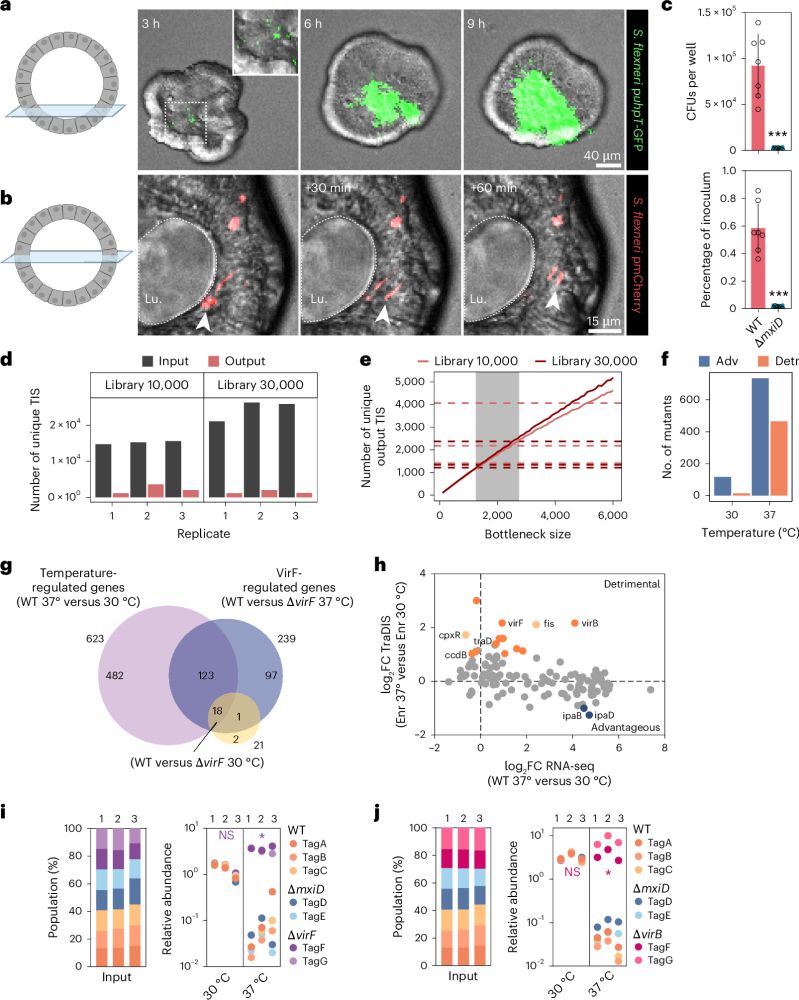
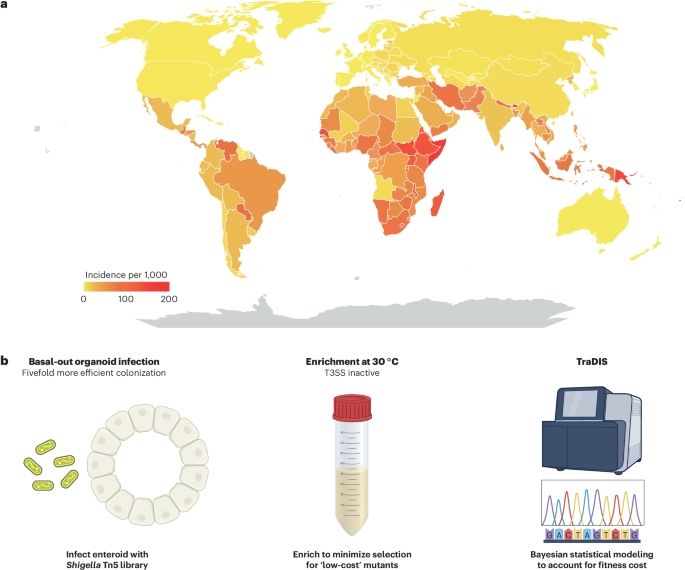
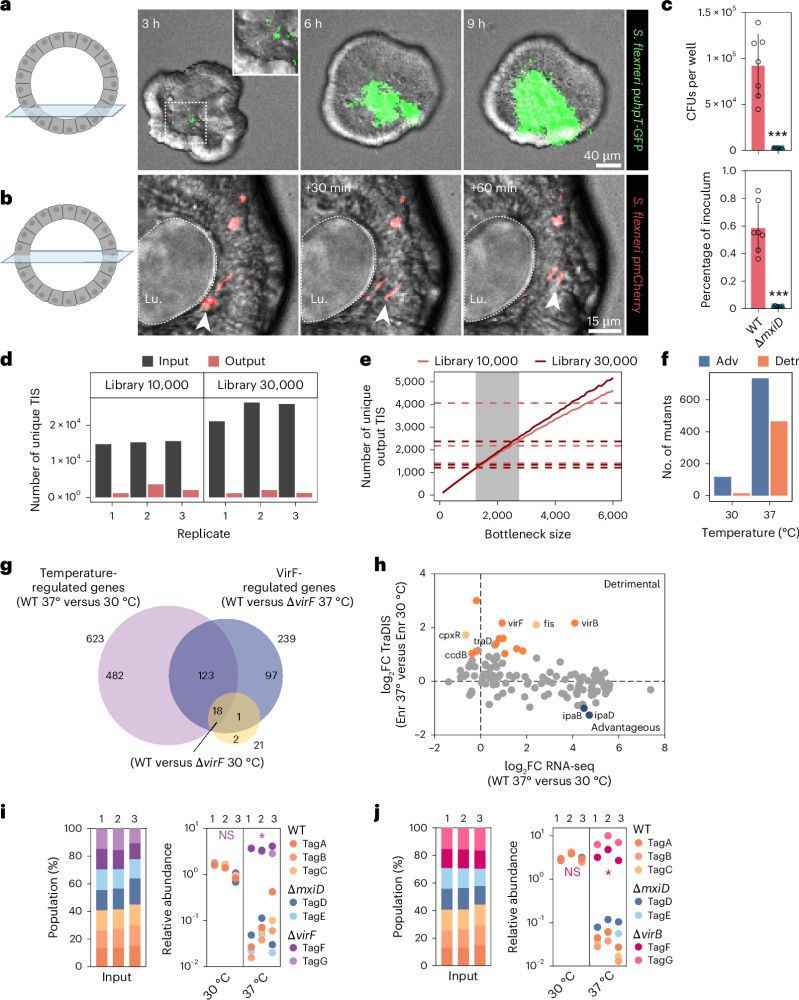
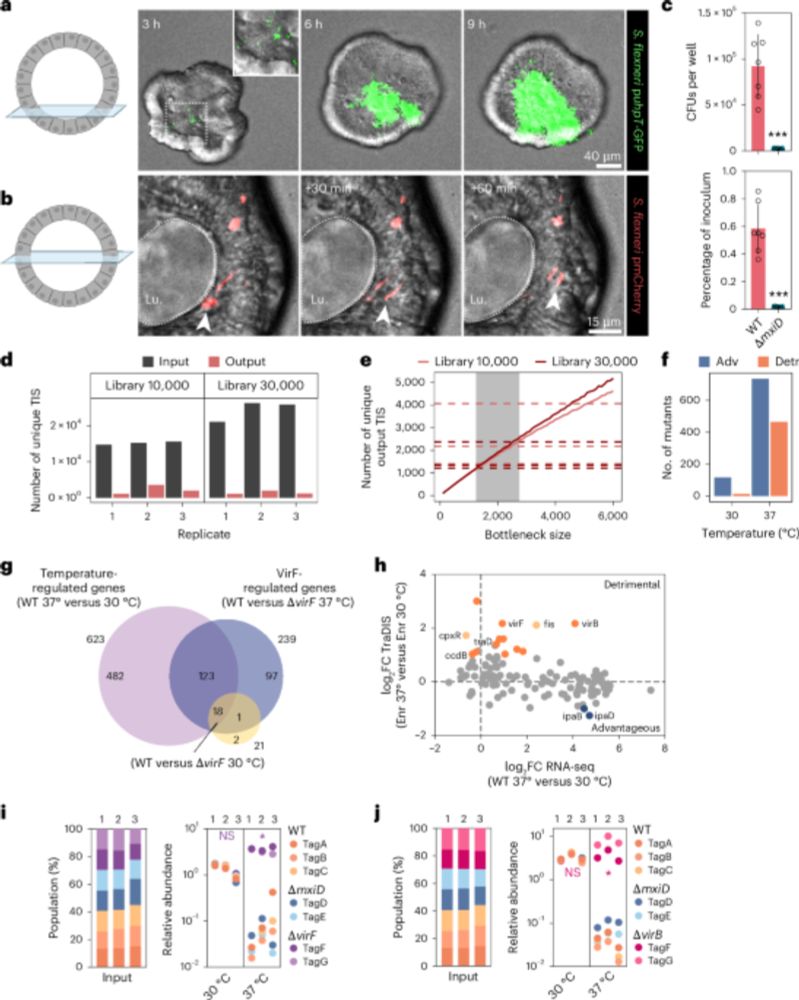
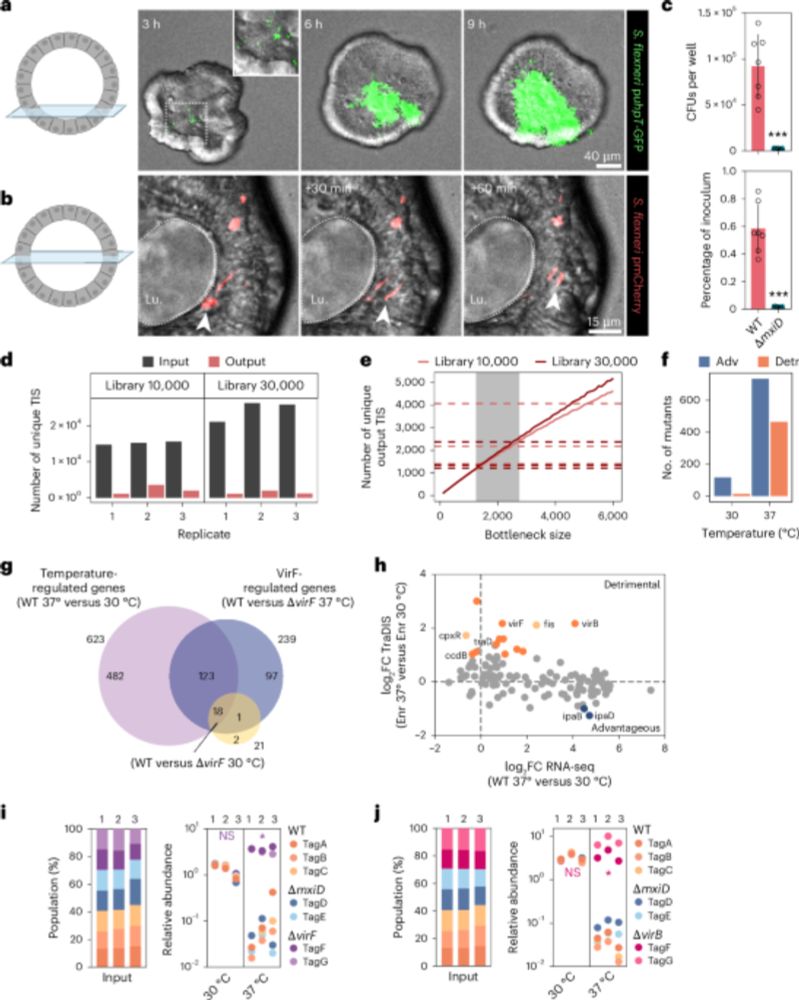
Combining a scalable human gut organoid model with TraDIS and statistical modeling, we mapped how Shigella colonizes the intestine at genome-wide resolution.

In short: pcnB ➝increased virulence-plasmid replication ➝ elevated T3SS expression ➝ enhanced invasion/spread within the intestinal epithelium.

In short: pcnB ➝increased virulence-plasmid replication ➝ elevated T3SS expression ➝ enhanced invasion/spread within the intestinal epithelium.
@sydneylmiles.bsky.social @katholt.bsky.social
🙏to transformative collaborative effort
www.nature.com/articles/s41...
@sydneylmiles.bsky.social @katholt.bsky.social
🙏to transformative collaborative effort
www.nature.com/articles/s41...

onlinelibrary.wiley.com/doi/10.1002/...

onlinelibrary.wiley.com/doi/10.1002/...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
Decoding Shigella’s Virulence Blueprint 🦠🧬
Combining organoid infection models with genome-wide mutagenesis allows mapping the full set of Shigella genes required for human gut colonization
www.nature.com/articles/s41...
@sellinlab.bsky.social @natgenet.nature.com
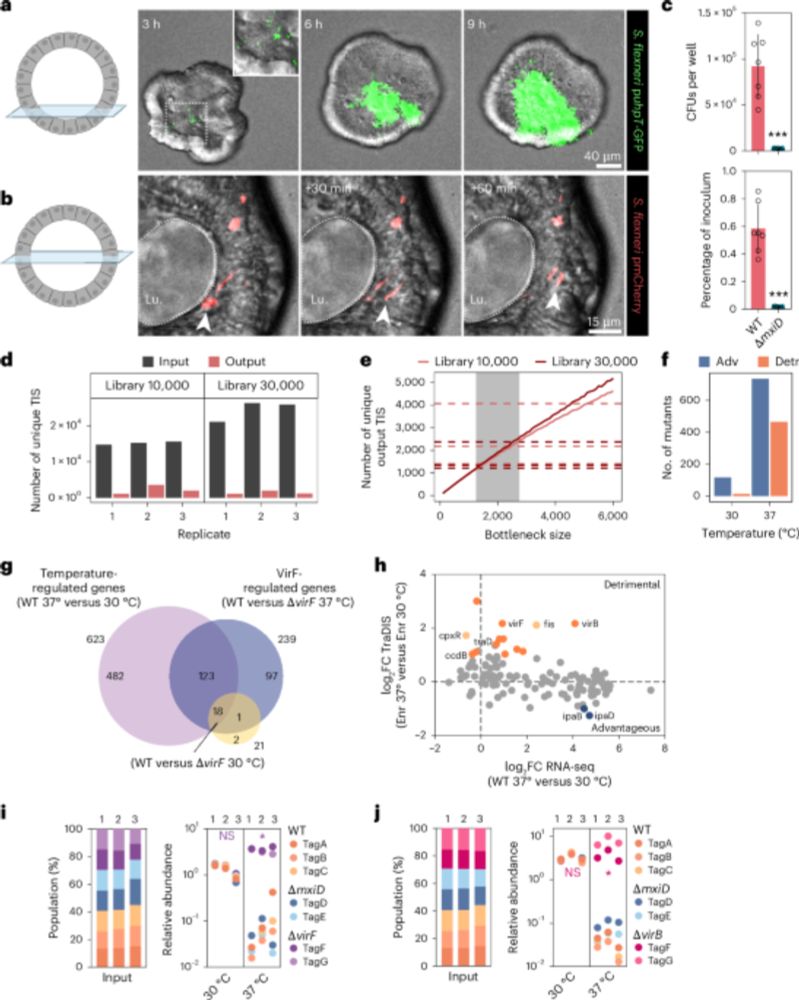
Decoding Shigella’s Virulence Blueprint 🦠🧬
Combining organoid infection models with genome-wide mutagenesis allows mapping the full set of Shigella genes required for human gut colonization
www.nature.com/articles/s41...
@sellinlab.bsky.social @natgenet.nature.com
Read our interview with study co-author — and UTM Biology prof — Lars Barquist at uoft.me/bI7.

Read our interview with study co-author — and UTM Biology prof — Lars Barquist at uoft.me/bI7.
Stop by poster 109 or just look for me, happy to chat and connect!
#Microbiology #Shigella #Organoids #TraDIS
📰A scalable gut epithelial organoid model reveals the genome-wide colonization landscape of a human-adapted pathogen.
By @mldm.bsky.social, @lbarquist.bsky.social, Mikael E. Sellin and colleagues.
⬇️
www.nature.com/articles/s41...

Stop by poster 109 or just look for me, happy to chat and connect!
#Microbiology #Shigella #Organoids #TraDIS
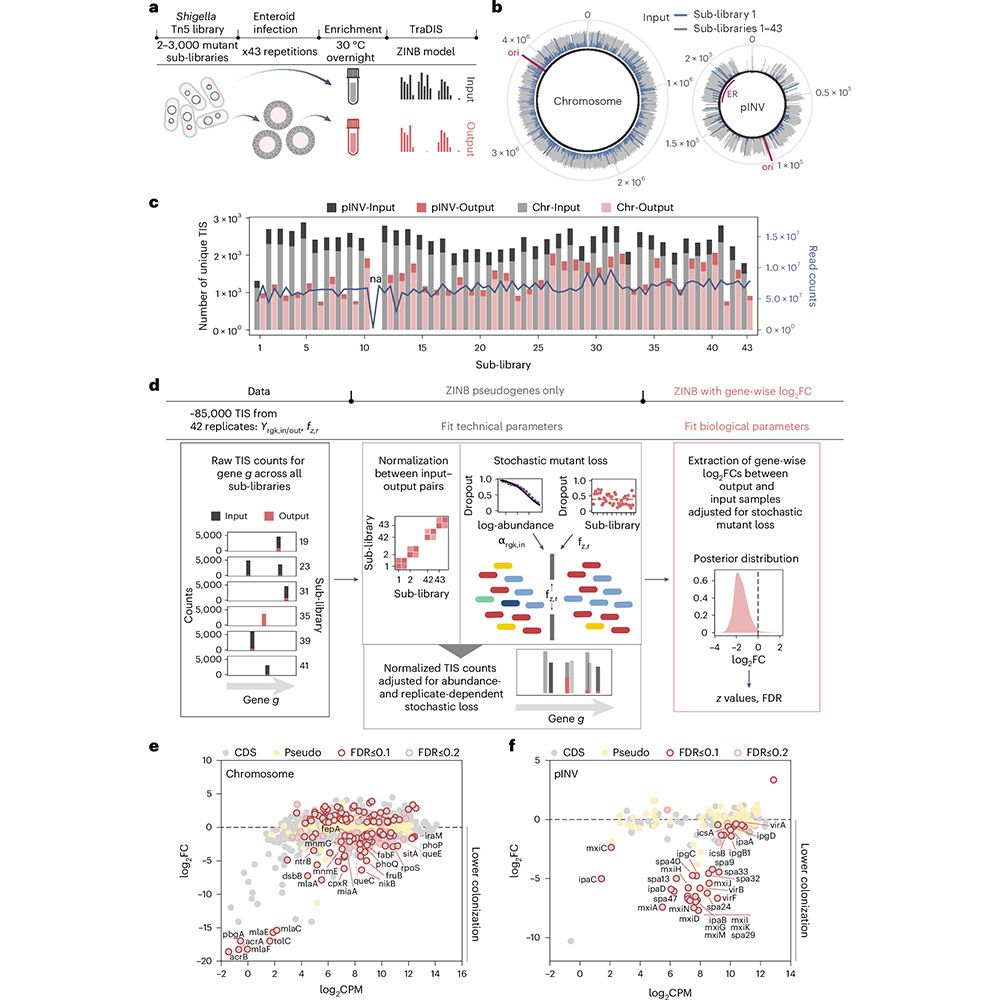
We followed up on our recent Nature Genetics paper mapping Shigella colonization in intestinal organoids with a deep dive into one of the screen hits: pcnB.
#Shigella #virulence #microsky
We followed up on our recent Nature Genetics paper mapping Shigella colonization in intestinal organoids with a deep dive into one of the screen hits: pcnB.
#Shigella #virulence #microsky
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Out @natgenet.nature.com:
rdcu.be/eqGgf
Out @natgenet.nature.com:
rdcu.be/eqGgf
www.nature.com/articles/s41...
www.nature.com/articles/s41...
@sellinlab.bsky.social (SciLifeLab & @uu.se) shows that cultured mini-organs can uncover how pathogens cause disease
youtu.be/xG_66ZGd_fk

@sellinlab.bsky.social (SciLifeLab & @uu.se) shows that cultured mini-organs can uncover how pathogens cause disease
youtu.be/xG_66ZGd_fk
www.nature.com/articles/s41...
www.nature.com/articles/s41...


