
www.biorxiv.org/content/10.1...
SMART-PTA for whole-genome+transcriptome on thousand of single cells from the normal human esophagus 🤯 Massively scaling up the power of scWGS to build deep phylogenies and chart somatic evolution from birth throughout life.
www.biorxiv.org/content/10.1...

SMART-PTA for whole-genome+transcriptome on thousand of single cells from the normal human esophagus 🤯 Massively scaling up the power of scWGS to build deep phylogenies and chart somatic evolution from birth throughout life.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.cell.com/cancer-cell/...

www.cell.com/cancer-cell/...
doi.org/10.1186/s130...

doi.org/10.1186/s130...

mathsdata2025.github.io
EPFL, Sept 1–5, 2025
Speakers:
Bach @bachfrancis.bsky.social
Bandeira
Mallat
Montanari
Peyré @gabrielpeyre.bsky.social
For PhD students & early-career researchers
Apply before May 15!
mathsdata2025.github.io
EPFL, Sept 1–5, 2025
Speakers:
Bach @bachfrancis.bsky.social
Bandeira
Mallat
Montanari
Peyré @gabrielpeyre.bsky.social
For PhD students & early-career researchers
Apply before May 15!
We present our new alignment framework TOAST www.biorxiv.org/content/10.1...
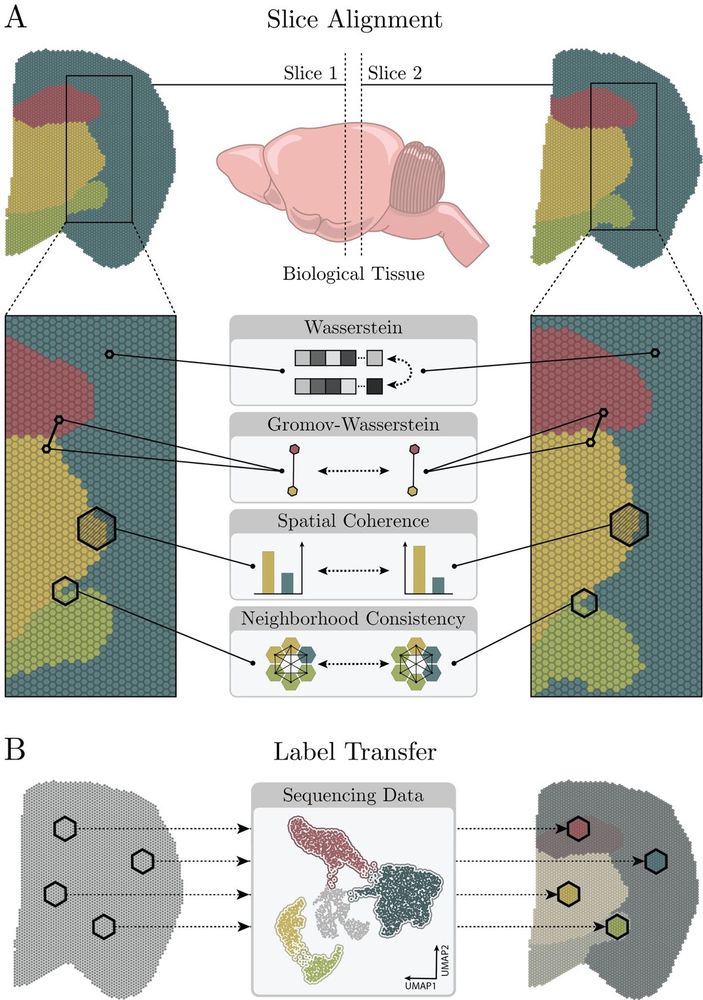
We present our new alignment framework TOAST www.biorxiv.org/content/10.1...
coursesandconferences.wellcomeconnectingscience.org/event/evolut...
Our brilliant faculty will cover evo&eco theory, math models, genomics, digital path, clinical trial design & more
coursesandconferences.wellcomeconnectingscience.org/event/evolut...
Our brilliant faculty will cover evo&eco theory, math models, genomics, digital path, clinical trial design & more
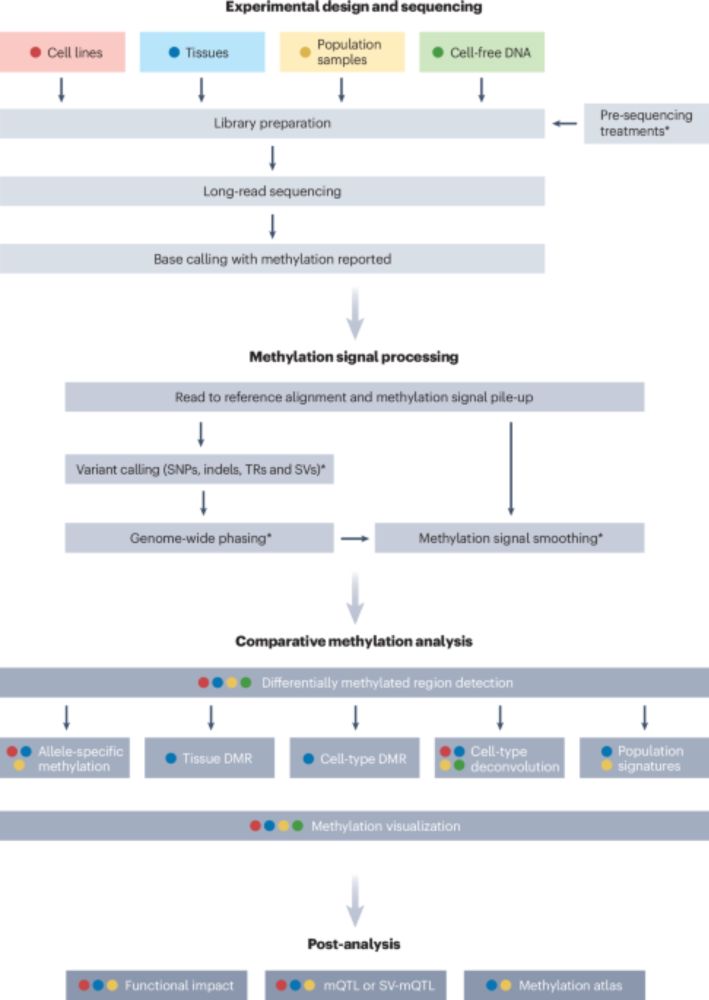
A detailed guide to assessing #genome-assembly based on #long-read sequencing data using Inspector. @natprot.bsky.social

A detailed guide to assessing #genome-assembly based on #long-read sequencing data using Inspector. @natprot.bsky.social
doi.org/10.1111/tan....

doi.org/10.1111/tan....
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
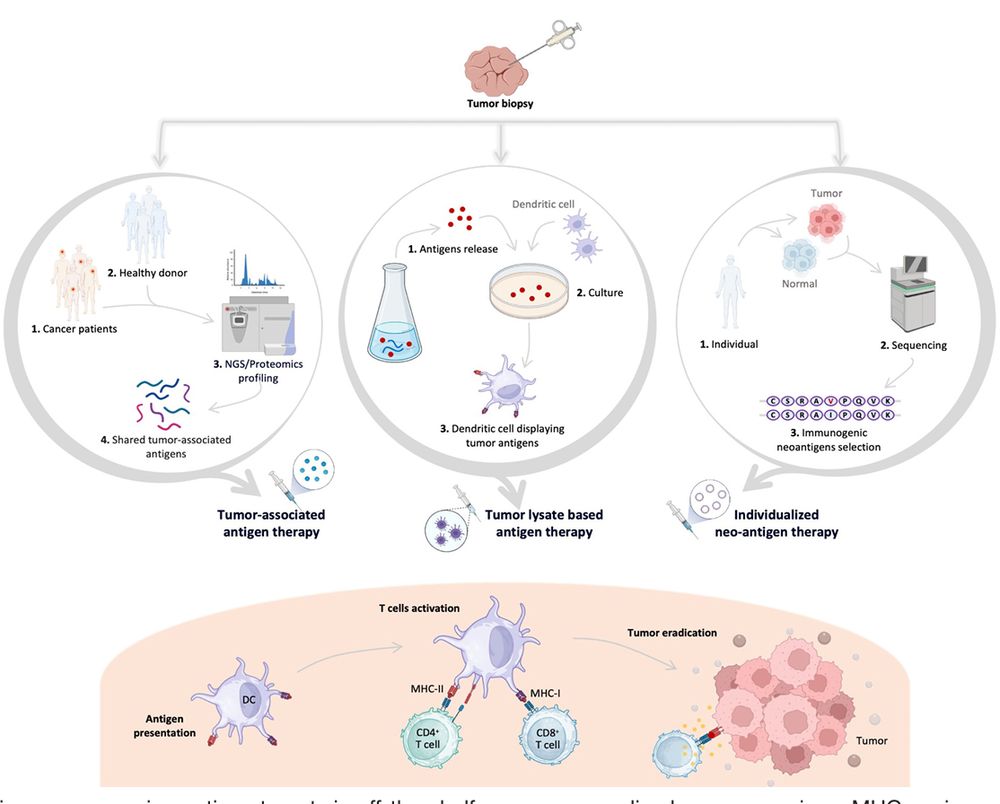
www.cell.com/cell/fullte...
www.cell.com/cell/fullte...
Why? Which ones?
We conducted an unbiased analysis of response biomarkers and found that hundreds of possible biomarkers collapse into five latent, orthogonal factors that underlie immunotherapy response.
www.nature.com/articles/s41...
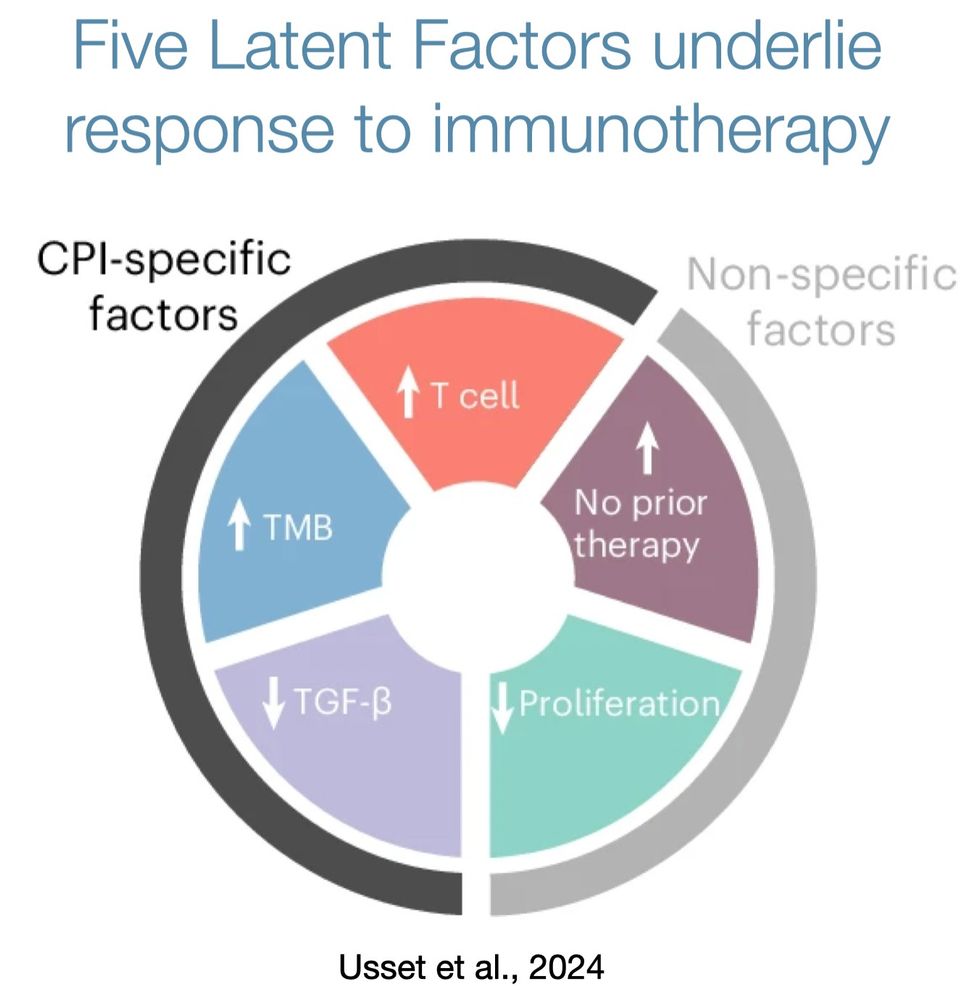
Why? Which ones?
We conducted an unbiased analysis of response biomarkers and found that hundreds of possible biomarkers collapse into five latent, orthogonal factors that underlie immunotherapy response.
www.nature.com/articles/s41...
Amazing to see even more improvement with bispecific antibody #immunotherapy presented in the #ASH24 Plenary session. #hematology #leukemia
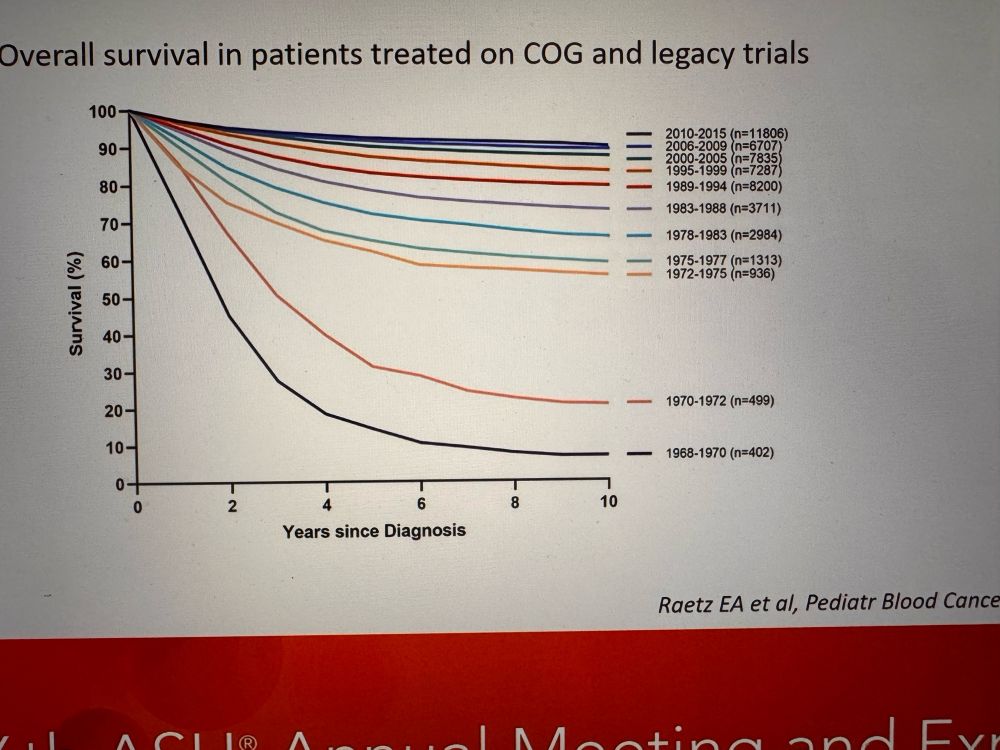
Amazing to see even more improvement with bispecific antibody #immunotherapy presented in the #ASH24 Plenary session. #hematology #leukemia


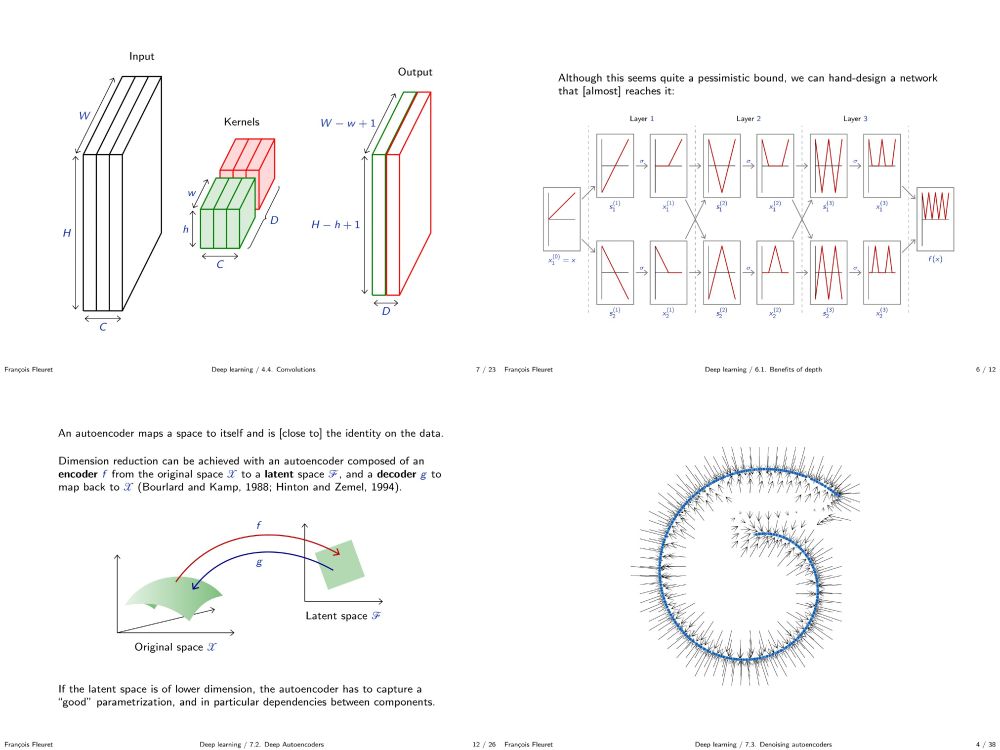
fleuret.org/dlc/
And my "Little Book of Deep Learning" is available as a phone-formatted pdf (nearing 700k downloads!)
fleuret.org/lbdl/
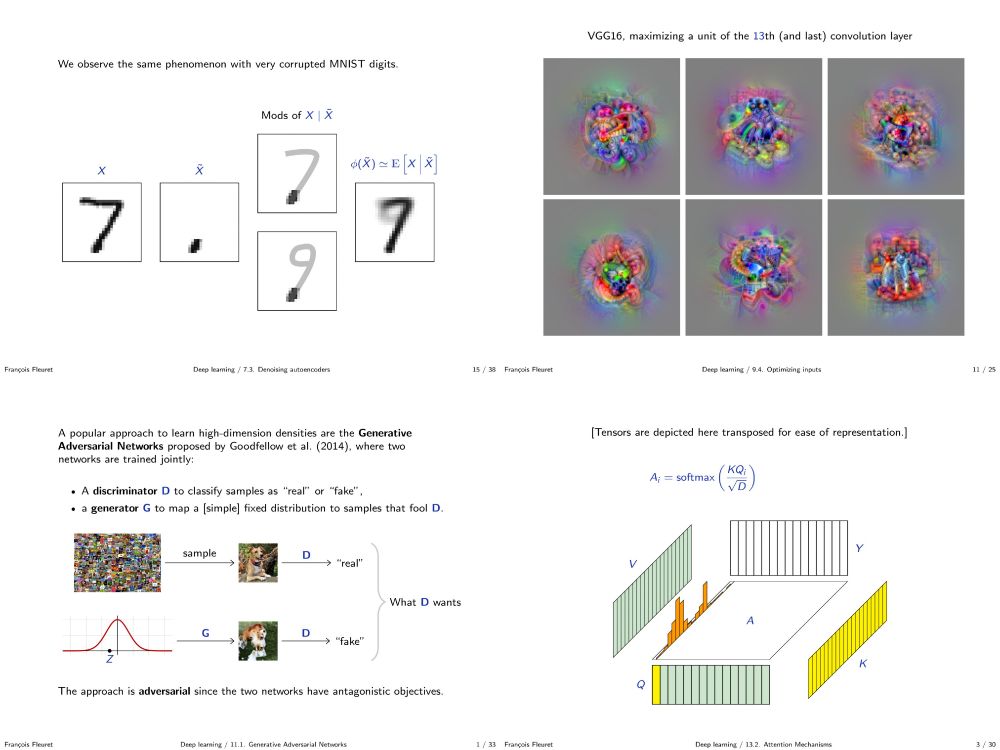
fleuret.org/dlc/
And my "Little Book of Deep Learning" is available as a phone-formatted pdf (nearing 700k downloads!)
fleuret.org/lbdl/



