
https://robinsonlabuzh.github.io/

(including interoperability with Python)
Paper: www.biorxiv.org/content/10.1...
Book: bioconductor.org/books/OSTA/
#Rstats


(including interoperability with Python)
Paper: www.biorxiv.org/content/10.1...
Book: bioconductor.org/books/OSTA/
#Rstats
Check it out, get in touch. We welcome any feedback, suggestions, wishes (& contributions).
It’s been a joy working with you @estellayixingdong.bsky.social!
Check it out, get in touch. We welcome any feedback, suggestions, wishes (& contributions).
It’s been a joy working with you @estellayixingdong.bsky.social!
By combining single-cell RNA sequencing and 3D imaging, we reveal striking structural and molecular differences between human and mouse dermal lymphatic vasculature, opening up new perspectives on #Lymphatics and #HumanBiology
By combining single-cell RNA sequencing and 3D imaging, we reveal striking structural and molecular differences between human and mouse dermal lymphatic vasculature, opening up new perspectives on #Lymphatics and #HumanBiology
⬇️
journals.plos.org/ploscompbiol...

⬇️
journals.plos.org/ploscompbiol...
https://www.biorxiv.org/content/10.1101/2025.11.06.686971v1
https://www.biorxiv.org/content/10.1101/2025.11.06.686971v1
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Expect 3 exciting days with the Bioconductor community, showcasing the latest advances in Bioconductor software and emerging technologies shaping computational biology.
👉 More details soon at eurobioc2026.bioconductor.org

Expect 3 exciting days with the Bioconductor community, showcasing the latest advances in Bioconductor software and emerging technologies shaping computational biology.
👉 More details soon at eurobioc2026.bioconductor.org
We introduce structure-based analysis of spatial omics data – an approach that focuses on multi-cellular anatomical structures rather than single cells.
We also present sosta to facilitate this type of analysis: bioconductor.org/packages/sos...
We introduce structure-based analysis of spatial omics data – an approach that focuses on multi-cellular anatomical structures rather than single cells.
We also present sosta to facilitate this type of analysis: bioconductor.org/packages/sos...


Day 1 was full of great talks, conversations, and that amazing Bioconductor community energy. Excited for what’s ahead today and tomorrow!
#EuroBioC2025 #Bioconductor #OpenScience #Community




Day 1 was full of great talks, conversations, and that amazing Bioconductor community energy. Excited for what’s ahead today and tomorrow!
#EuroBioC2025 #Bioconductor #OpenScience #Community
academic.oup.com/nar/article/...

academic.oup.com/nar/article/...
We discuss the broad range of exploratory spatial statistics options for spatial Omics technologies and show relevant use cases.
arxiv.org/abs/2412.01561

We discuss the broad range of exploratory spatial statistics options for spatial Omics technologies and show relevant use cases.
arxiv.org/abs/2412.01561
More in our preprint: www.biorxiv.org/content/10.1...

More in our preprint: www.biorxiv.org/content/10.1...
@pacbio.bsky.social @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...
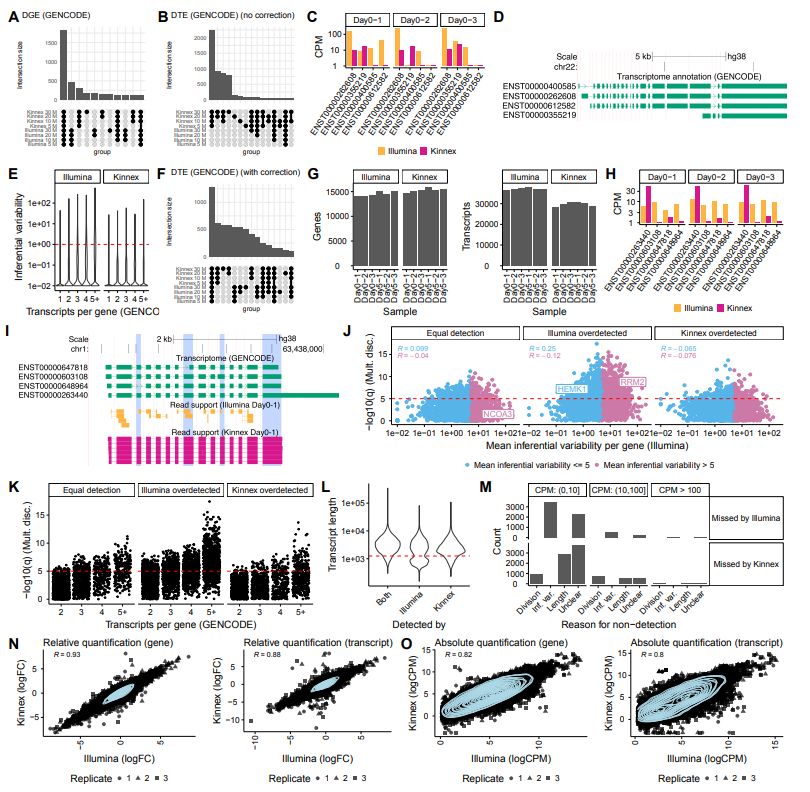
@pacbio.bsky.social @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...
⭐ gold-standard training data through curated databases
🔗 connected information through knowledge representation
💪 trustworthy evaluation through benchmarking.
See how in the #SIBProfile 2025: issuu.com/sibswissinst...
⭐ gold-standard training data through curated databases
🔗 connected information through knowledge representation
💪 trustworthy evaluation through benchmarking.
See how in the #SIBProfile 2025: issuu.com/sibswissinst...
* #muscat doi.org/10.1038/s414... @helucro.bsky.social
* pbDD doi.org/10.1101/2023... @davi1893.bsky.social
* #dreamlet doi.org/10.1101/2023... @panosroussos.bsky.social
* #distinct doi.org/10.1214/22-A... @markrobinsonca.bsky.social
youtu.be/TB9dfKPfns8

* #muscat doi.org/10.1038/s414... @helucro.bsky.social
* pbDD doi.org/10.1101/2023... @davi1893.bsky.social
* #dreamlet doi.org/10.1101/2023... @panosroussos.bsky.social
* #distinct doi.org/10.1214/22-A... @markrobinsonca.bsky.social
youtu.be/TB9dfKPfns8
www.biomedcentral.com/collections/...
www.biomedcentral.com/collections/...

⬇️
journals.plos.org/ploscompbiol...
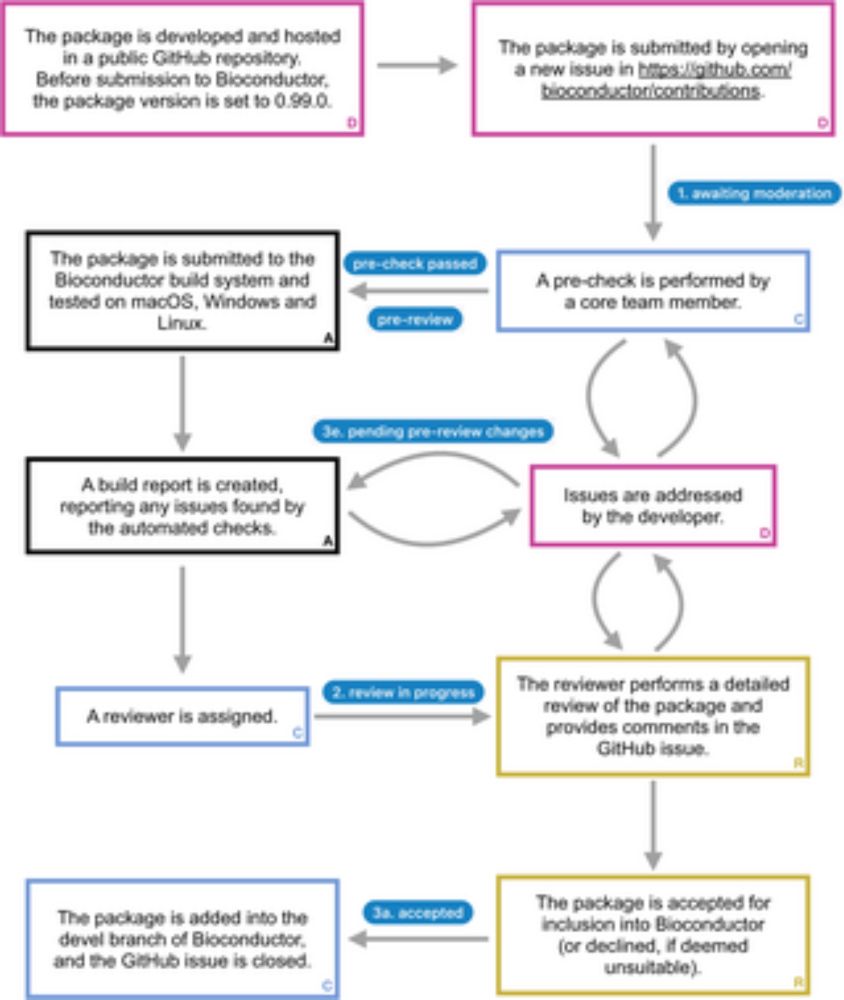
⬇️
journals.plos.org/ploscompbiol...
sites.google.com/view/ascona2...
There are few registration slots left, so if you are interested to join us, get in touch.
sites.google.com/view/ascona2...
There are few registration slots left, so if you are interested to join us, get in touch.

