Now published in Nature Nanotechnology! 🤩
@natnano.nature.com
www.nature.com/articles/s41...
www.nature.com/articles/s41...
rdcu.be/eQVxt

www.nature.com/articles/s41...
rdcu.be/eQVxt
A special poster prize committee elected three winners:
Alara Kiris, @lumasullo.bsky.social and @wanluzhang.bsky.social. Congratulations!
Read on to learn about their research: s.embl.org/ees25-09post...

A special poster prize committee elected three winners:
Alara Kiris, @lumasullo.bsky.social and @wanluzhang.bsky.social. Congratulations!
Read on to learn about their research: s.embl.org/ees25-09post...
- Focus Issue on #biosensing,
- DNA moiré superlattices,
- Sugars at Ångström-resolution,
- Solid-state #nanopores,
- Non-aqueous Li #batteries, -
- Neuromorphic vision,
- Peptide #hydrogels,
- Deep learning for #LNPs and more...
www.nature.com/nnano/volume...

- Focus Issue on #biosensing,
- DNA moiré superlattices,
- Sugars at Ångström-resolution,
- Solid-state #nanopores,
- Non-aqueous Li #batteries, -
- Neuromorphic vision,
- Peptide #hydrogels,
- Deep learning for #LNPs and more...
www.nature.com/nnano/volume...
www.nature.com/nnano/volume...

www.nature.com/nnano/volume...
Excited to share that I’ve been awarded a Wellcome Trust Early Career Award to establish my research as a Group Leader in the Glycosciences Program at the Institute of Biology and Experimental Medicine (IBYME) in Buenos Aires, Argentina, beginning in early 2026.
Excited to share that I’ve been awarded a Wellcome Trust Early Career Award to establish my research as a Group Leader in the Glycosciences Program at the Institute of Biology and Experimental Medicine (IBYME) in Buenos Aires, Argentina, beginning in early 2026.




us06web.zoom.us/webinar/regi...
#maxplanck #mpimr #colloquium #medicalresearch #sciencetalks #science

us06web.zoom.us/webinar/regi...
#maxplanck #mpimr #colloquium #medicalresearch #sciencetalks #science
www.nature.com/articles/s41...

www.nature.com/articles/s41...
Now published in Nature Nanotechnology! 🤩
@natnano.nature.com
www.nature.com/articles/s41...
Now published in Nature Nanotechnology! 🤩
@natnano.nature.com
www.nature.com/articles/s41...
Ångström-resolution imaging of cell-surface glycans.
The molecular organization of sugars in the native #glycocalyx has been resolved at 9 ångström using bioorthogonal metabolic labeling and #superresolution imaging of DNA barcodes.
#Glycotime
www.nature.com/articles/s41...
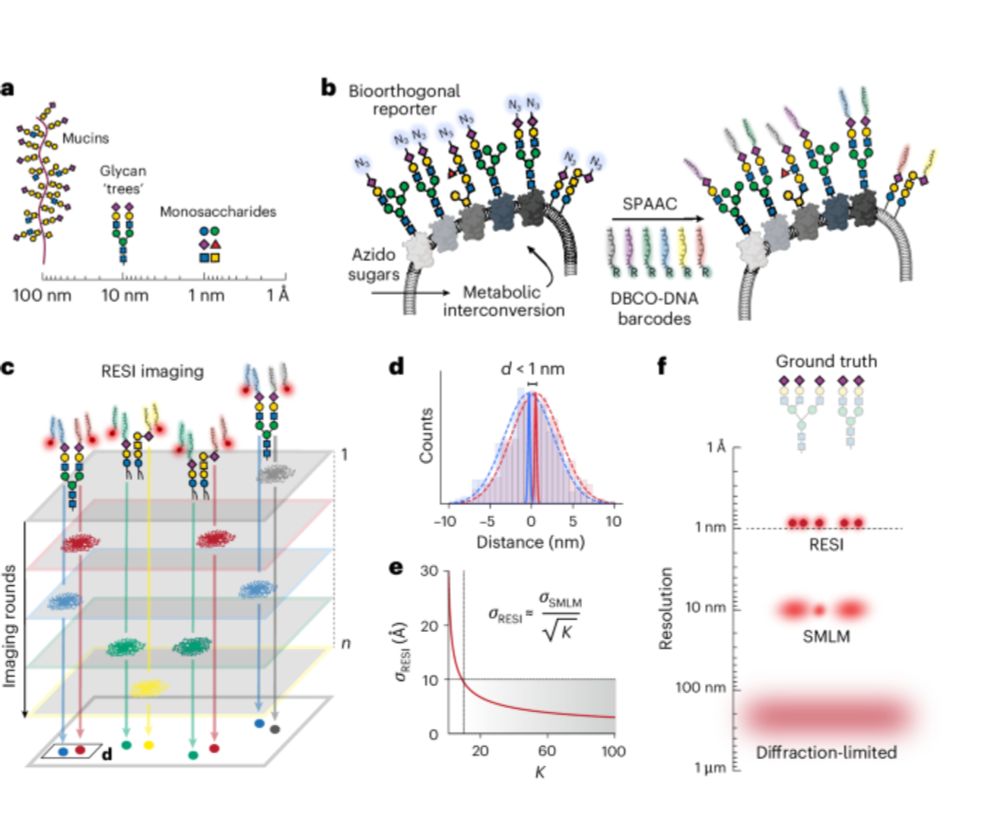
Ångström-resolution imaging of cell-surface glycans.
The molecular organization of sugars in the native #glycocalyx has been resolved at 9 ångström using bioorthogonal metabolic labeling and #superresolution imaging of DNA barcodes.
#Glycotime
www.nature.com/articles/s41...
Read the full story here: www.nature.com/articles/s41...
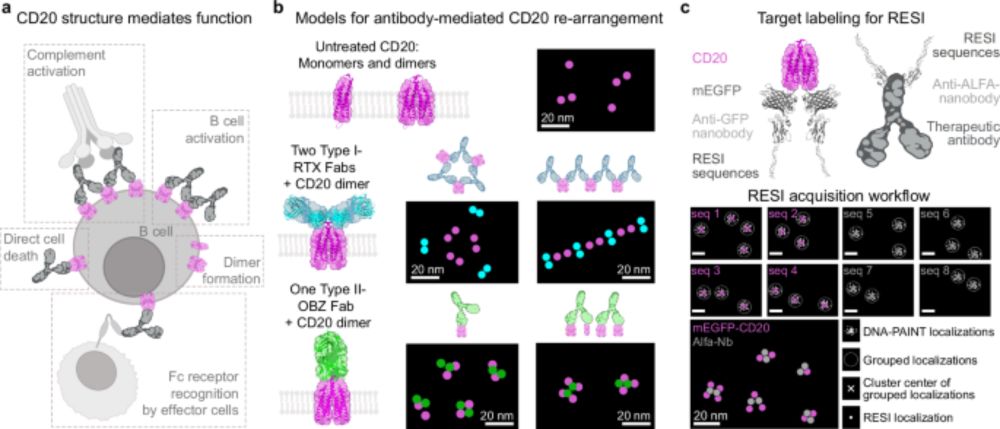
Read the full story here: www.nature.com/articles/s41...
🧬🔬 YES: with #DNA_PAINT + Spinning Disk Confocal + Optical Photon Reassignment (#SDC-OPR)!
📄 www.nature.com/articles/s41... @natcomms.nature.com!
🧵👇
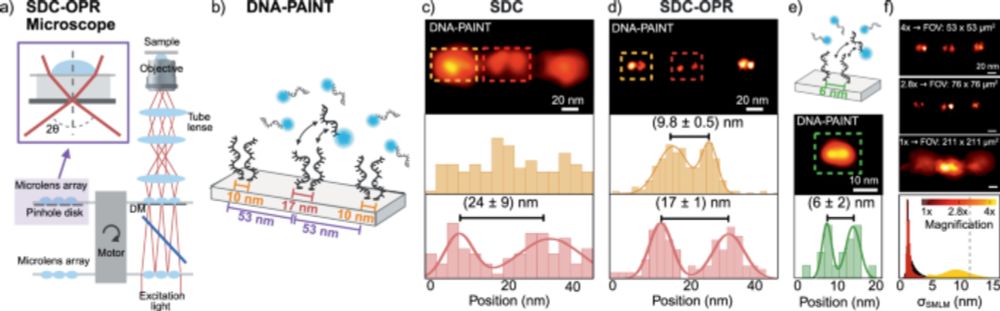
🧬🔬 YES: with #DNA_PAINT + Spinning Disk Confocal + Optical Photon Reassignment (#SDC-OPR)!
📄 www.nature.com/articles/s41... @natcomms.nature.com!
🧵👇
📄 bit.ly/4jTHfes
📄 bit.ly/4jTHfes
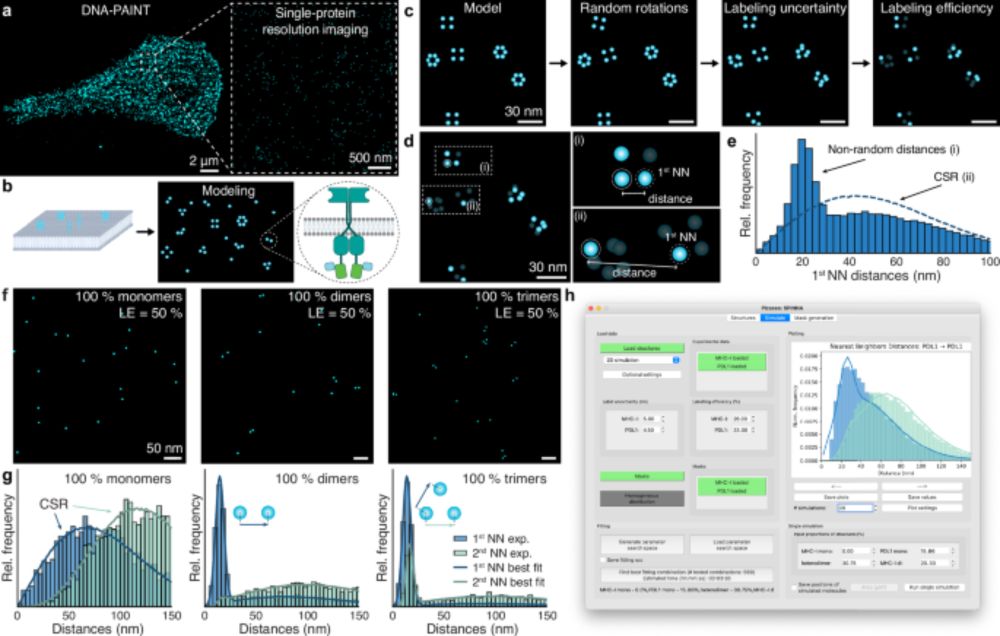
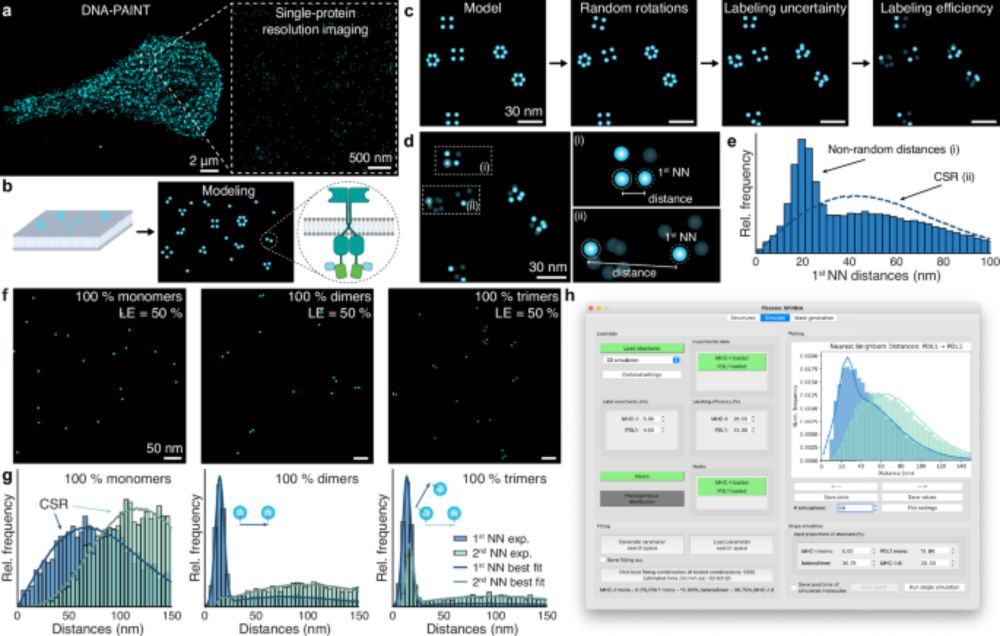
We can directly quantify stoichiometry and oligomerization from super-res (DNA-PAINT, RESI) images!! 🧬🎨
We are excited to present our latest work published in @natcomms.nature.com
www.nature.com/articles/s41...
We can directly quantify stoichiometry and oligomerization from super-res (DNA-PAINT, RESI) images!! 🧬🎨
We are excited to present our latest work published in @natcomms.nature.com
www.nature.com/articles/s41...

We are excited to present our latest work published in @natcomms.nature.com
www.nature.com/articles/s41...

Prof. Xi and his team are doing amazing work on various type of SIM and other imaging modalities!
Thanks so much for the warm welcome, the stimulating discussions, and the campus tour!!




Prof. Xi and his team are doing amazing work on various type of SIM and other imaging modalities!
Thanks so much for the warm welcome, the stimulating discussions, and the campus tour!!
They’re doing top-notch research, including awesome interferometric single-molecule localization methods (ROSE)
thanks so much for the invitation, warm welcome and hospitality!! 😍🤩



They’re doing top-notch research, including awesome interferometric single-molecule localization methods (ROSE)
thanks so much for the invitation, warm welcome and hospitality!! 😍🤩
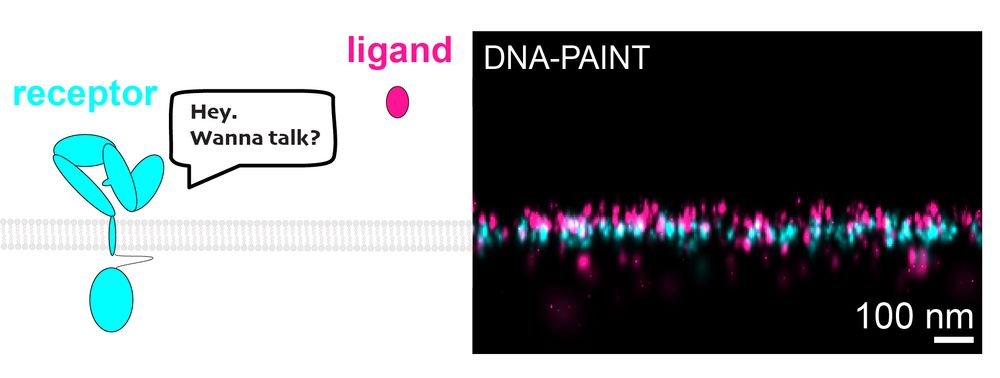
Ever wonder how cells "talk"? It starts when ligands bind to receptors on cell surfaces. We have cracked the challenge of imaging small ligands on cell surfaces. #DNAPAINT #celltalk 💬
doi.org/10.1002/smtd...
Ever wonder how cells "talk"? It starts when ligands bind to receptors on cell surfaces. We have cracked the challenge of imaging small ligands on cell surfaces. #DNAPAINT #celltalk 💬
doi.org/10.1002/smtd...
Project led by my extremely talented colleagues Monique Honsa, Larissa Heinze and Isabelle Pachmayr
Very happy to have contributed to this beautiful work! 🤩
Ever wonder how cells "talk"? It starts when ligands bind to receptors on cell surfaces. We have cracked the challenge of imaging small ligands on cell surfaces. #DNAPAINT #celltalk 💬
doi.org/10.1002/smtd...

Project led by my extremely talented colleagues Monique Honsa, Larissa Heinze and Isabelle Pachmayr
Very happy to have contributed to this beautiful work! 🤩



