Excited to share our discovery of the RAD51 filament cap, built by a newly identified RAD51 paralog complex 🔬🧬
‼️ Excited to share our new paper out now in @science.org ‼️
We describe a new tetrameric RAD51 paralog complex – XRCC3-RAD51C-RAD51D-XRCC2 – which caps the end of RAD51 filaments.
Link: www.science.org/doi/epdf/10....
Thread ⬇️ (1/8)
We describe a new tetrameric RAD51 paralog complex – XRCC3-RAD51C-RAD51D-XRCC2 – which caps the end of RAD51 filaments.
Link: www.science.org/doi/epdf/10....
Thread ⬇️ (1/8)
November 7, 2025 at 10:22 AM
Excited to share our discovery of the RAD51 filament cap, built by a newly identified RAD51 paralog complex 🔬🧬
Reposted by Luke Greenhough
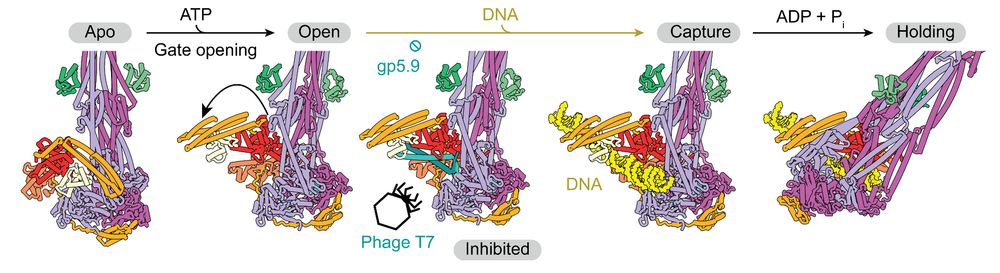
How do SMC complexes capture DNA? 🧪
Led by Frank Bürmann, Jan Löwe’s & Mark Dillingham’s groups have identified the directional loading mechanism the bacterial SMC complex MukBEF uses to capture & ingest DNA, & found a phage inhibitor to this pathway.
Read more: tinyurl.com/4s8nrrkk
#LMBResearch
Led by Frank Bürmann, Jan Löwe’s & Mark Dillingham’s groups have identified the directional loading mechanism the bacterial SMC complex MukBEF uses to capture & ingest DNA, & found a phage inhibitor to this pathway.
Read more: tinyurl.com/4s8nrrkk
#LMBResearch
March 31, 2025 at 2:40 PM
How do SMC complexes capture DNA? 🧪
Led by Frank Bürmann, Jan Löwe’s & Mark Dillingham’s groups have identified the directional loading mechanism the bacterial SMC complex MukBEF uses to capture & ingest DNA, & found a phage inhibitor to this pathway.
Read more: tinyurl.com/4s8nrrkk
#LMBResearch
Led by Frank Bürmann, Jan Löwe’s & Mark Dillingham’s groups have identified the directional loading mechanism the bacterial SMC complex MukBEF uses to capture & ingest DNA, & found a phage inhibitor to this pathway.
Read more: tinyurl.com/4s8nrrkk
#LMBResearch
Reposted by Luke Greenhough
Over five years of work has gone into our new preprint! The #cryoEM structure of a human RAD51 filament caught at the point of strand exchange 🧬
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Structural mechanism of strand exchange by the RAD51 filament.
Homologous Recombination (HR) preserves genomic stability by repairing double-strand DNA breaks and ensuring efficient DNA replication. Central to HR is the strand-exchange reaction taking place within the three-stranded synapsis wherein a RAD51 nucleoprotein filament binds to a donor DNA. Here we present the cryoEM structure of a displacement loop of human RAD51 that captures the synaptic state when the filament has become tightly bound to the donor DNA. The structure elucidates the mechanism of strand exchange by RAD51, including the filament engagement with the donor DNA, the strand invasion and pairing with the complementary sequence of the donor DNA, the capture of the non-complementary strand and the polarity of the strand-exchange reaction. Our findings provide fundamental mechanistic insights into the biochemical reaction of eukaryotic HR. ### Competing Interest Statement The authors have declared no competing interest.
www.biorxiv.org
March 27, 2025 at 3:12 PM
Over five years of work has gone into our new preprint! The #cryoEM structure of a human RAD51 filament caught at the point of strand exchange 🧬
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Luke Greenhough
🚀 New Paper Alert! Our latest @natrevmcb.bsky.social explores how nuclear and genome organization drive DNA double-strand break repair! 🧬
📖 Free access: rdcu.be/edViW
Please 🔄 ❤️
📖 Free access: rdcu.be/edViW
Please 🔄 ❤️
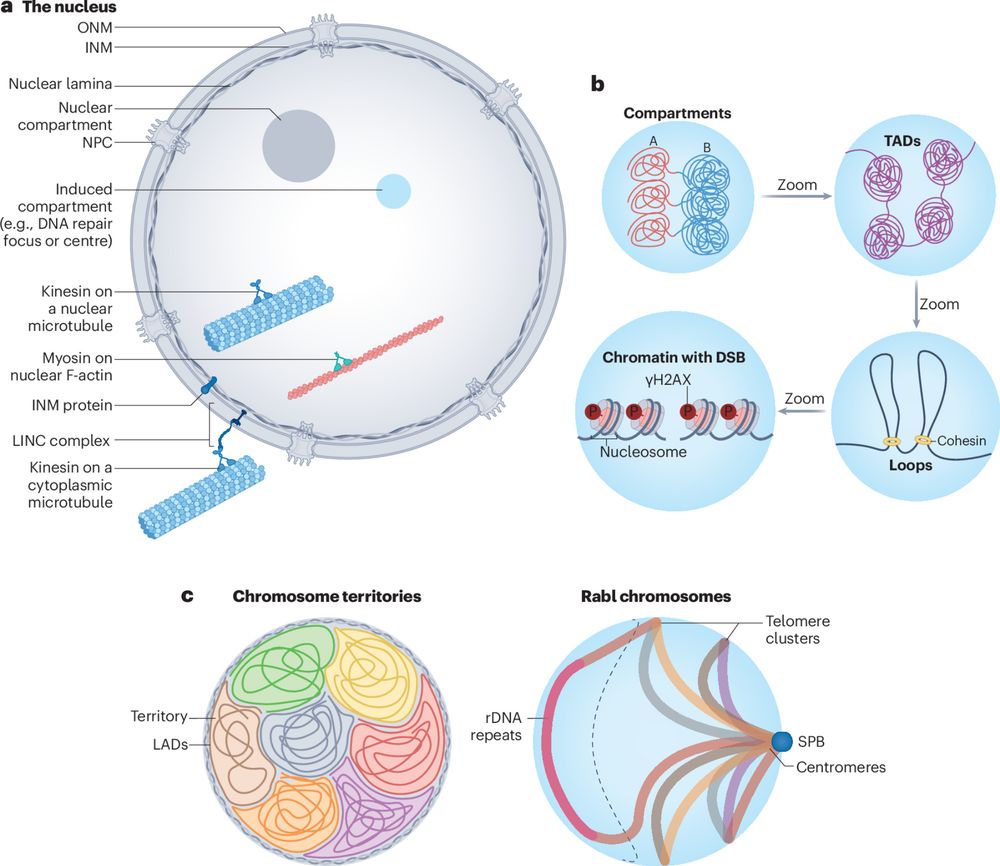
Nuclear and genome dynamics underlying DNA double-strand break repair
Nature Reviews Molecular Cell Biology - Changes in nuclear and genome organization promote the repair of DNA double-strand breaks and genome stability. Processes that are involved include the...
rdcu.be
March 18, 2025 at 1:49 PM
🚀 New Paper Alert! Our latest @natrevmcb.bsky.social explores how nuclear and genome organization drive DNA double-strand break repair! 🧬
📖 Free access: rdcu.be/edViW
Please 🔄 ❤️
📖 Free access: rdcu.be/edViW
Please 🔄 ❤️
Reposted by Luke Greenhough
Check out our review on DNA end resection! With Raphael Ceccaldi, www.nature.com/articles/s41...

March 25, 2025 at 4:00 PM
Check out our review on DNA end resection! With Raphael Ceccaldi, www.nature.com/articles/s41...
Been excited for this for a while! Excited to chat about all things paralog related
📅 Our next Chromosome Biology Club is next week!
Join us on March 25th at the Biocenter for exciting talks from:
◉ Johanna Kliche (Nilsson group, @nilssonlab.bsky.social)
◉ Oline Pade Jensen (Lisby group)
◉ Luke Greenhough (Steve West group, @xrnchew.bsky.social)
Mark your calendars! #CBC
Join us on March 25th at the Biocenter for exciting talks from:
◉ Johanna Kliche (Nilsson group, @nilssonlab.bsky.social)
◉ Oline Pade Jensen (Lisby group)
◉ Luke Greenhough (Steve West group, @xrnchew.bsky.social)
Mark your calendars! #CBC

March 25, 2025 at 7:21 AM
Been excited for this for a while! Excited to chat about all things paralog related
Reposted by Luke Greenhough
Excited to see the final published form of this amazing @drewberry.bsky.social animation of DNA repair go live! Watch your favourite friends BRCA1/2, RPA, RAD51, Exo1, BLM, PCNA, MRN, primase and DNA polymerase perform their recombination magic. Cool educational & scientific tool for our community.
Delighted to publish my new molecular animation:
DNA Break Repair by Homologous Recombination
youtu.be/Xe-83tBcxhs
DNA Break Repair by Homologous Recombination
youtu.be/Xe-83tBcxhs
DNA Break Repair by Homologous Recombination (2024) Drew Berry wehi.tv
YouTube video by WEHImovies
youtu.be
December 4, 2024 at 1:46 AM
Excited to see the final published form of this amazing @drewberry.bsky.social animation of DNA repair go live! Watch your favourite friends BRCA1/2, RPA, RAD51, Exo1, BLM, PCNA, MRN, primase and DNA polymerase perform their recombination magic. Cool educational & scientific tool for our community.
Reposted by Luke Greenhough
Very excited to share that my thesis work is out in Molecular Cell! We trained a Structure and Omics informed Classifier (SPOC) to score binary AlphaFold multimer (AF-M) predictions by structural quality and consistency with experimental omics datasets. www.cell.com/molecular-ce...
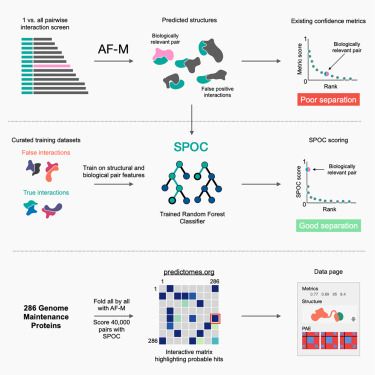
Predictomes, a classifier-curated database of AlphaFold-modeled protein-protein interactions
Schmid and Walter train a classifier that discerns functionally relevant structure
predictions in proteome-wide protein-protein interaction (PPI) screens using AlphaFold-Multimer,
and they use this co...
www.cell.com
February 26, 2025 at 6:14 PM
Very excited to share that my thesis work is out in Molecular Cell! We trained a Structure and Omics informed Classifier (SPOC) to score binary AlphaFold multimer (AF-M) predictions by structural quality and consistency with experimental omics datasets. www.cell.com/molecular-ce...
Reposted by Luke Greenhough
Thank you to Roger Greenberg and Hilda Pickett for organising a fantastic 2025 Mammalian DNA Repair GRC in Ventura, California.
Looking forward to the next time.
Looking forward to the next time.


February 7, 2025 at 12:37 AM
Thank you to Roger Greenberg and Hilda Pickett for organising a fantastic 2025 Mammalian DNA Repair GRC in Ventura, California.
Looking forward to the next time.
Looking forward to the next time.

