
Structural biology & DNA repair.
When not freezing grids I'm baking, climbing, hiking or taking photos.
Have a paper people should see? See what our Editors look for: buff.ly/rr68suW
Have a paper people should see? See what our Editors look for: buff.ly/rr68suW
buff.ly/ofD02RD
buff.ly/ofD02RD
buff.ly/Pz8MI7W
buff.ly/Pz8MI7W
➡️ www.path.cam.ac.uk/news/pacific...

➡️ www.path.cam.ac.uk/news/pacific...
elifesciences.org/reviewed-pre...

elifesciences.org/reviewed-pre...
📰 Press release: elifesciences.org/for-the-pres...
📄 Full article: elifesciences.org/reviewed-pre...
Proud of the team #DNADamage #CryoEM #CambridgeUniversity
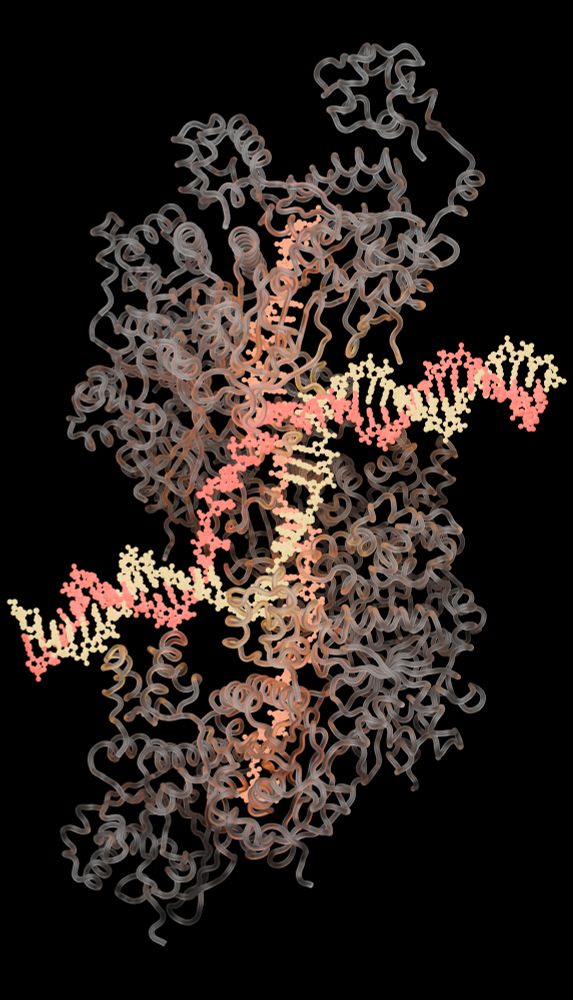
📰 Press release: elifesciences.org/for-the-pres...
📄 Full article: elifesciences.org/reviewed-pre...
Proud of the team #DNADamage #CryoEM #CambridgeUniversity
www.nature.com/articles/s41...
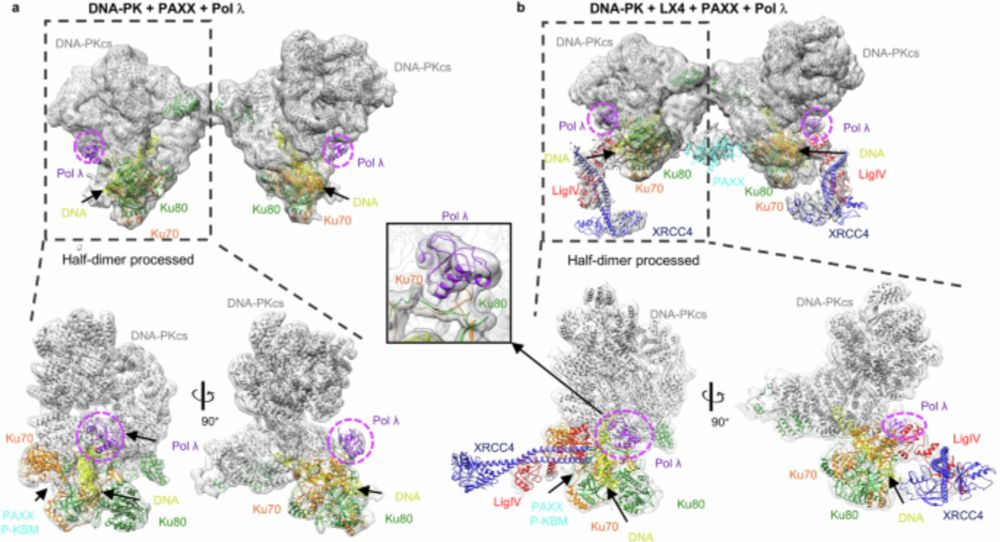
www.nature.com/articles/s41...
www.path.cam.ac.uk/research/fac...
www.path.cam.ac.uk/research/fac...



You'll find a tutorial on how to reconstruct tomograms, pick particles and do subtomogram averaging, using different software!
Hope it will be useful !


You'll find a tutorial on how to reconstruct tomograms, pick particles and do subtomogram averaging, using different software!
Hope it will be useful !
@marinalusic.bsky.social, Turoňová lab, @hummerlab.bsky.social @becklab.bsky.social @mpibp.bsky.social
🔗 Preprint here tinyurl.com/3a74uanv
@marinalusic.bsky.social, Turoňová lab, @hummerlab.bsky.social @becklab.bsky.social @mpibp.bsky.social
🔗 Preprint here tinyurl.com/3a74uanv
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

