
www.medrxiv.org/content/10.1...
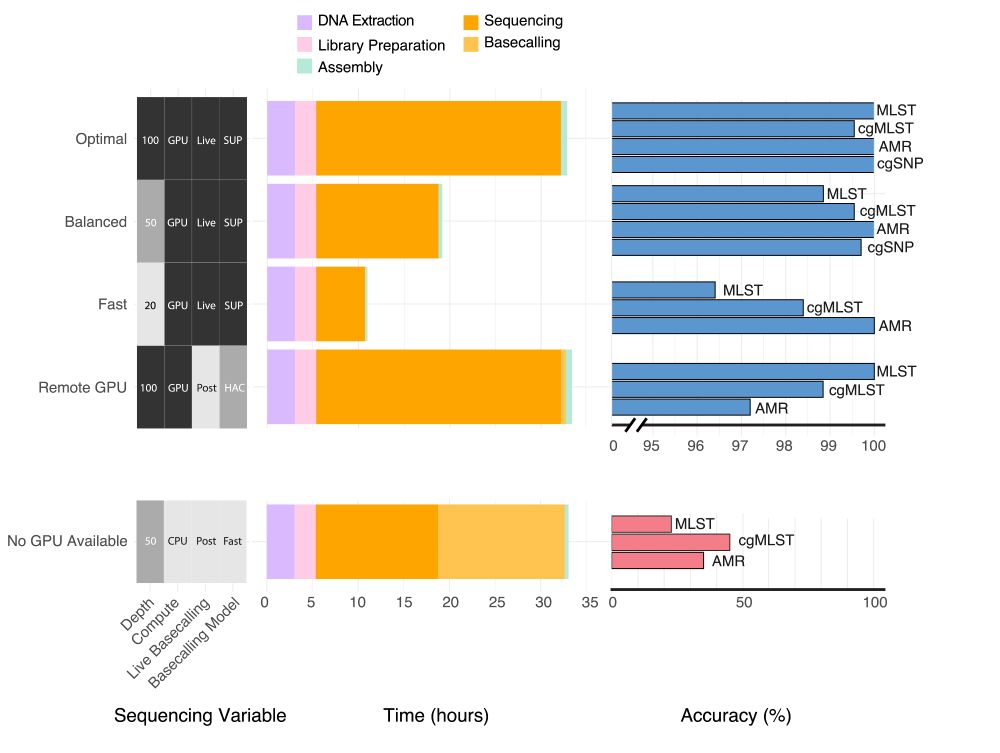
www.medrxiv.org/content/10.1...
My phage annotation tool, Phynteny, finally has a preprint and a brand new version powered by a cool AI transformer architecture and protein language models! #phagesky
www.biorxiv.org/content/10.1...

My phage annotation tool, Phynteny, finally has a preprint and a brand new version powered by a cool AI transformer architecture and protein language models! #phagesky
www.biorxiv.org/content/10.1...
Need a quick explainer on the new names? Check out my blog post: tinyurl.com/y8yb3rbb (+link to full review article)
#MicroSky @klebnet.bsky.social @tomstantonmicro.bsky.social
Need a quick explainer on the new names? Check out my blog post: tinyurl.com/y8yb3rbb (+link to full review article)
#MicroSky @klebnet.bsky.social @tomstantonmicro.bsky.social
Thanks @kjolley.bsky.social!
Also like to acknowledge that the Acineto dbs were developed and maintained by Johanna Kenyon and team at Griffith Uni.

Thanks @kjolley.bsky.social!
Also like to acknowledge that the Acineto dbs were developed and maintained by Johanna Kenyon and team at Griffith Uni.
#MicroSeq2025

#MicroSeq2025
#MicroSky #IDSky #GlobalHealth 🧬💻🧪 (1/3)
www.medrxiv.org/content/10.1...

#MicroSky #IDSky #GlobalHealth 🧬💻🧪 (1/3)
www.medrxiv.org/content/10.1...
doi.org/10.1101/2025...

Want to collaborate globally to advance Klebs genomic epi? ✅
Please join us via the KlebNET-GSP Epidemiology consortium ⬇️
#microsky #AMR
We aim to build a public metadata repository; systematic risk framework for global genomic surveillance; and genomic epi reviews for high-impact #Klebsiella clones.
Join us here:
klebnet.org/klebnet-gsp-...
#ABPHM25
Want to collaborate globally to advance Klebs genomic epi? ✅
Please join us via the KlebNET-GSP Epidemiology consortium ⬇️
#microsky #AMR
🎉🎉🎉
@aussocmic.bsky.social
🎉🎉🎉
@aussocmic.bsky.social
🦠🧫
Big thanks to coauthors @kelwyres.bsky.social, @katholt.bsky.social, @genomarit.bsky.social and Iren Löhr.
Here's what we did to improve in silico antigen typing 👇🧵
www.biorxiv.org/content/10.1...

🦠🧫
Some years ago many people were compiling 𝐥𝐢𝐬𝐭𝐬 𝐨𝐟 𝐰𝐨𝐦𝐞𝐧 𝐢𝐧 𝐬𝐜𝐢𝐞𝐧𝐜𝐞 as an effort to break biases when looking for reviewers and collaborators...
Is there anything like that for Bioinformatics and Genomics?
#WomenInSTEM
Some years ago many people were compiling 𝐥𝐢𝐬𝐭𝐬 𝐨𝐟 𝐰𝐨𝐦𝐞𝐧 𝐢𝐧 𝐬𝐜𝐢𝐞𝐧𝐜𝐞 as an effort to break biases when looking for reviewers and collaborators...
Is there anything like that for Bioinformatics and Genomics?
#WomenInSTEM
Bonus points if not E. coli. #microsky

All info here: www.who.int/news-room/ar...

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

K. pneumoniae contributes to "pathway to death" in almost a quarter of infants between months 1 and 59 months of life.
#KLEBS2024

K. pneumoniae contributes to "pathway to death" in almost a quarter of infants between months 1 and 59 months of life.
#KLEBS2024
#MicroSky
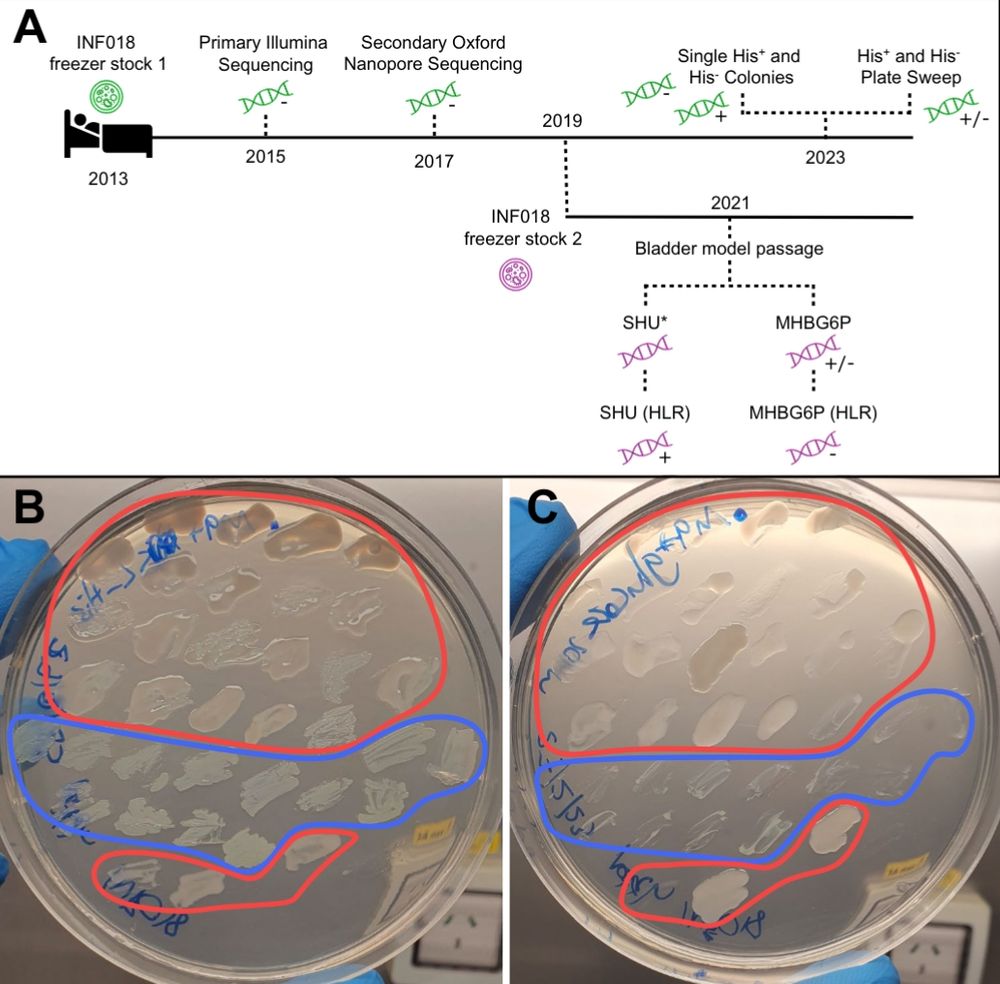
#MicroSky
400 in person and 400 online participants from 71 countries...... Wow!

400 in person and 400 online participants from 71 countries...... Wow!
Looking forward to catching up with great collaborators and friends, and making new connections with Klebs aficionados. 🎉🎉🎉
Looking forward to catching up with great collaborators and friends, and making new connections with Klebs aficionados. 🎉🎉🎉

