www.imperial.ac.uk/news/271753/...

www.imperial.ac.uk/news/271753/...
Our kick-off meeting for the Klebsiella Seminar Series just wrapped up. Stay tuned for more!
@lauraamike.bsky.social @olayarendueles.bsky.social @caityholmes.bsky.social @tomstantonmicro.bsky.social @juanvalenciabacca.bsky.social WenWen Low & Jay V.
Our kick-off meeting for the Klebsiella Seminar Series just wrapped up. Stay tuned for more!
@lauraamike.bsky.social @olayarendueles.bsky.social @caityholmes.bsky.social @tomstantonmicro.bsky.social @juanvalenciabacca.bsky.social WenWen Low & Jay V.
Topics include Kleb diversity, lineages, AMR, hypervirulence, how to use Kaptive & Kleborate for typing, and more!
klebnet.org/2025/11/18/k...
Topics include Kleb diversity, lineages, AMR, hypervirulence, how to use Kaptive & Kleborate for typing, and more!
klebnet.org/2025/11/18/k...

Need a quick explainer on the new names? Check out my blog post: tinyurl.com/y8yb3rbb (+link to full review article)
#MicroSky @klebnet.bsky.social @tomstantonmicro.bsky.social
Need a quick explainer on the new names? Check out my blog post: tinyurl.com/y8yb3rbb (+link to full review article)
#MicroSky @klebnet.bsky.social @tomstantonmicro.bsky.social
We aim to build a public metadata repository; systematic risk framework for global genomic surveillance; and genomic epi reviews for high-impact #Klebsiella clones.
Join us here:
klebnet.org/klebnet-gsp-...
#ABPHM25
We aim to build a public metadata repository; systematic risk framework for global genomic surveillance; and genomic epi reviews for high-impact #Klebsiella clones.
Join us here:
klebnet.org/klebnet-gsp-...
#ABPHM25
📌The rapid detection of a neonatal unit outbreak of a wild-type Klebsiella variicola using decentralized Oxford Nanopore sequencing
doi.org/10.1186/s137...
@nanoporetech.com
🖥️🧬💻
#AcademicSky
#Microsky
🧪🧫🦠

📌The rapid detection of a neonatal unit outbreak of a wild-type Klebsiella variicola using decentralized Oxford Nanopore sequencing
doi.org/10.1186/s137...
@nanoporetech.com
🖥️🧬💻
#AcademicSky
#Microsky
🧪🧫🦠

Big thanks to coauthors @kelwyres.bsky.social, @katholt.bsky.social, @genomarit.bsky.social and Iren Löhr.
Here's what we did to improve in silico antigen typing 👇🧵
www.biorxiv.org/content/10.1...

Big thanks to coauthors @kelwyres.bsky.social, @katholt.bsky.social, @genomarit.bsky.social and Iren Löhr.
Here's what we did to improve in silico antigen typing 👇🧵
www.biorxiv.org/content/10.1...
Preprint now here: www.biorxiv.org/content/10.1...
Including a case study on nosocomial transmission of vancomycin resistant Enterococcus faecium
Preprint now here: www.biorxiv.org/content/10.1...
Including a case study on nosocomial transmission of vancomycin resistant Enterococcus faecium
#Klebsiella #KLEBS2024
join.slack.com/t/klebclub/s...
#Klebsiella #KLEBS2024
join.slack.com/t/klebclub/s...
You can also share to help us gain visibility
#MicroSky
go.bsky.app/EdereoU
You can also share to help us gain visibility
#MicroSky
go.bsky.app/EdereoU

2009 was a big year for Klebs!
- First genome published
- First description of ST258 KPC-producing clone
pubmed.ncbi.nlm.nih.gov/19218573/
- First description of NDM-1 beta-lactamase
pubmed.ncbi.nlm.nih.gov/19770275/
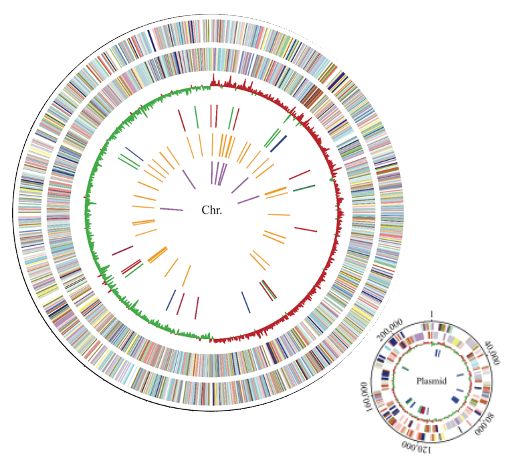
2009 was a big year for Klebs!
- First genome published
- First description of ST258 KPC-producing clone
pubmed.ncbi.nlm.nih.gov/19218573/
- First description of NDM-1 beta-lactamase
pubmed.ncbi.nlm.nih.gov/19770275/

#MicroSky
#MicroSky

