Tom Stanton
@tomstantonmicro.bsky.social
Microbiologist. Lead developer of #Kaptive + bonafide #Klebsiella nerd. Post-doc in the Wyres Lab @AlfredMonash_ID.
Kaptive 3 is also much (much) faster than Kaptive 2, taking ~1 second per assembly 🏎️💨
This means that if you don't have a fancy HPC, then don't worry! You can still analyse thousands of your own assemblies on your laptop in a reasonable time! 💻
This means that if you don't have a fancy HPC, then don't worry! You can still analyse thousands of your own assemblies on your laptop in a reasonable time! 💻

February 9, 2025 at 3:19 AM
Kaptive 3 is also much (much) faster than Kaptive 2, taking ~1 second per assembly 🏎️💨
This means that if you don't have a fancy HPC, then don't worry! You can still analyse thousands of your own assemblies on your laptop in a reasonable time! 💻
This means that if you don't have a fancy HPC, then don't worry! You can still analyse thousands of your own assemblies on your laptop in a reasonable time! 💻
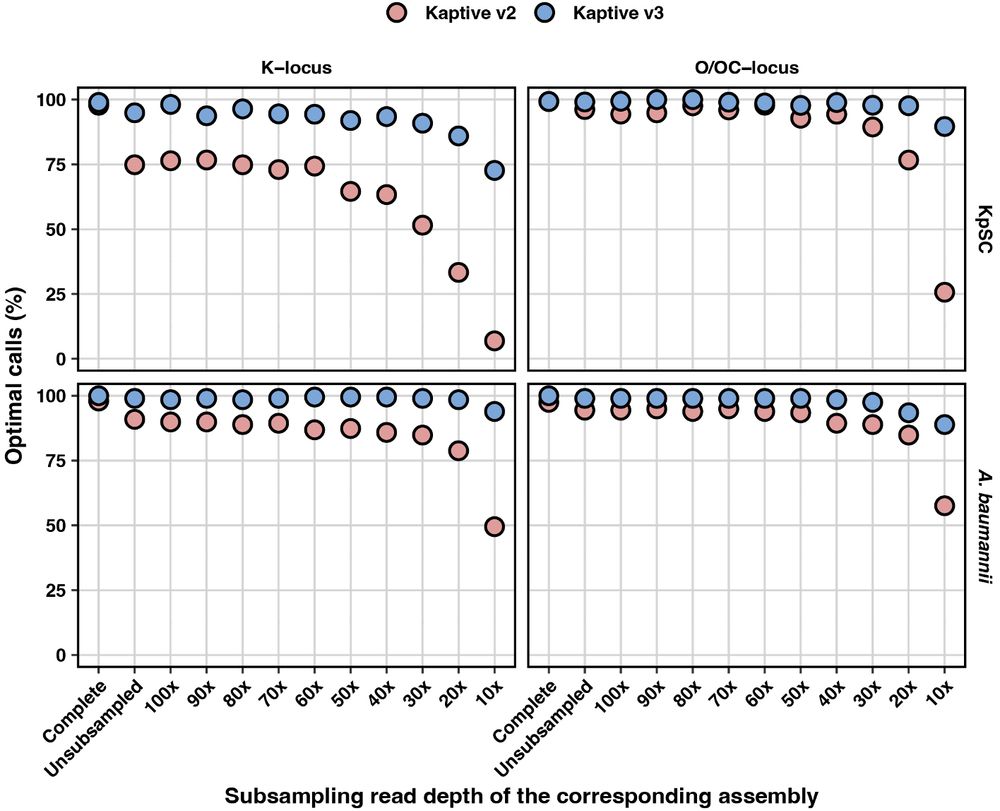
We then subsampled the corresponding short reads at decreasing depths and created sets of increasingly awful draft assemblies with loci broken over contigs and lots of genes missing.
Kaptive 3 was much more sensitive than Kaptive 2, and maintained accuracy even when the assemblies were awful! 💩
Kaptive 3 was much more sensitive than Kaptive 2, and maintained accuracy even when the assemblies were awful! 💩
February 9, 2025 at 3:19 AM
We then subsampled the corresponding short reads at decreasing depths and created sets of increasingly awful draft assemblies with loci broken over contigs and lots of genes missing.
Kaptive 3 was much more sensitive than Kaptive 2, and maintained accuracy even when the assemblies were awful! 💩
Kaptive 3 was much more sensitive than Kaptive 2, and maintained accuracy even when the assemblies were awful! 💩
So enter Kaptive 3, a complete overhaul of Kaptive with a new algorithm designed to handle fragmented loci.
We also refactored (and simplified) the confidence score to be more sensitive for broken loci and missing genes, allowing more Kaptive data to be used when the assembly may not be complete 💯
We also refactored (and simplified) the confidence score to be more sensitive for broken loci and missing genes, allowing more Kaptive data to be used when the assembly may not be complete 💯

February 9, 2025 at 3:19 AM
So enter Kaptive 3, a complete overhaul of Kaptive with a new algorithm designed to handle fragmented loci.
We also refactored (and simplified) the confidence score to be more sensitive for broken loci and missing genes, allowing more Kaptive data to be used when the assembly may not be complete 💯
We also refactored (and simplified) the confidence score to be more sensitive for broken loci and missing genes, allowing more Kaptive data to be used when the assembly may not be complete 💯
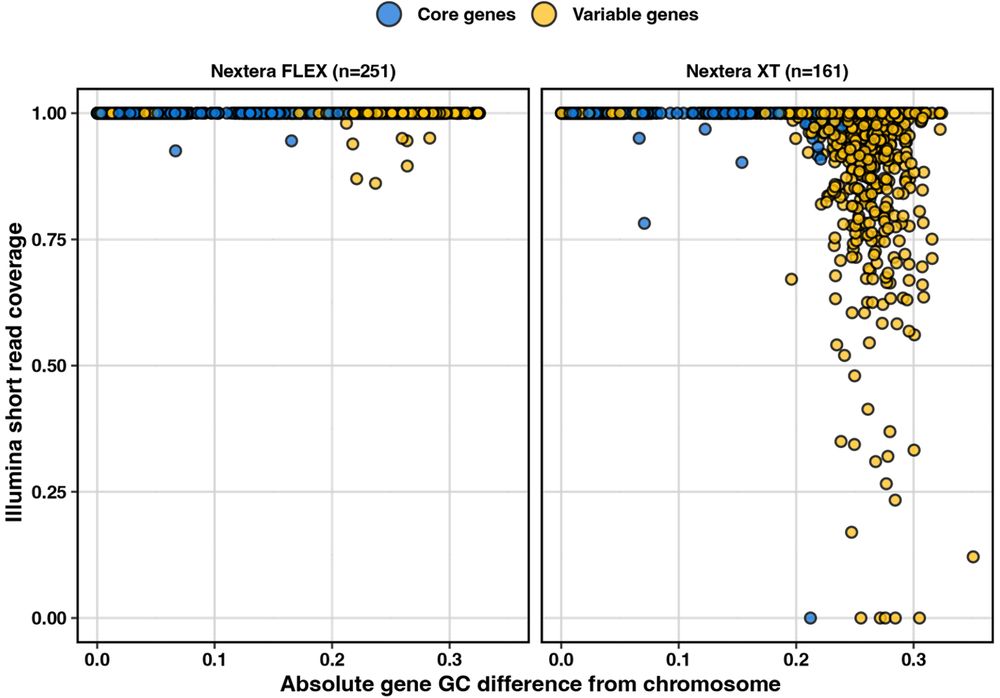
These genes have a very low GC compared to the rest of the Klebsiella chromosome, so we wondered if this was affecting how this part of the genome gets sequenced.
Turns out, these genes show decreased sequencing coverage when reads are prepped with Nextera XT, but not so much with Nextera Flex 🤯
Turns out, these genes show decreased sequencing coverage when reads are prepped with Nextera XT, but not so much with Nextera Flex 🤯
February 9, 2025 at 3:19 AM
These genes have a very low GC compared to the rest of the Klebsiella chromosome, so we wondered if this was affecting how this part of the genome gets sequenced.
Turns out, these genes show decreased sequencing coverage when reads are prepped with Nextera XT, but not so much with Nextera Flex 🤯
Turns out, these genes show decreased sequencing coverage when reads are prepped with Nextera XT, but not so much with Nextera Flex 🤯

