Tom Stanton
@tomstantonmicro.bsky.social
Microbiologist. Lead developer of #Kaptive + bonafide #Klebsiella nerd. Post-doc in the Wyres Lab @AlfredMonash_ID.
Reposted by Tom Stanton
Always a pleasure to organize this with @caityholmes.bsky.social @lauraamike.bsky.social Jay Vornhagen, Wen wen low, & this year joining us @tomstantonmicro.bsky.social & Juan Valencia.
April 1, 2025 at 8:30 AM
Always a pleasure to organize this with @caityholmes.bsky.social @lauraamike.bsky.social Jay Vornhagen, Wen wen low, & this year joining us @tomstantonmicro.bsky.social & Juan Valencia.
Janitor saves the day again!
www.rdocumentation.org/packages/jan...
www.rdocumentation.org/packages/jan...
excel_numeric_to_date function - RDocumentation
<p>Converts numbers like <code>42370</code> into date values like
<code>2016-01-01</code>.</p>
<p>Defaults to the modern Excel date encoding system. However, Excel for Mac
2008 and earlier Mac version...
www.rdocumentation.org
February 19, 2025 at 4:05 AM
Janitor saves the day again!
www.rdocumentation.org/packages/jan...
www.rdocumentation.org/packages/jan...
Lastly, we'd like to thank YOU, the Kaptive community, for guiding development, spotting bugs and collaborating with us!
But this is just the beginning, we have lots of exciting things in store for the future of Kaptive to make in silico serotyping even better!
#kaptive #klebsiella #acinetobacter
But this is just the beginning, we have lots of exciting things in store for the future of Kaptive to make in silico serotyping even better!
#kaptive #klebsiella #acinetobacter
February 9, 2025 at 3:19 AM
Lastly, we'd like to thank YOU, the Kaptive community, for guiding development, spotting bugs and collaborating with us!
But this is just the beginning, we have lots of exciting things in store for the future of Kaptive to make in silico serotyping even better!
#kaptive #klebsiella #acinetobacter
But this is just the beginning, we have lots of exciting things in store for the future of Kaptive to make in silico serotyping even better!
#kaptive #klebsiella #acinetobacter
Kaptive 3 is now integrated within Kaptive-Web (kaptive-web.erc.monash.edu), PathogenWatch (pathogen.watch), the new Kleborate 3 framework (github.com/klebgenomics...) and Bactopia (bactopia.github.io/latest/).
Remember to cite us if you use Kaptive for your results, and watch out for "Untypeable"!
Remember to cite us if you use Kaptive for your results, and watch out for "Untypeable"!
Pathogenwatch
A global platform for genomic surveillance.
pathogen.watch
February 9, 2025 at 3:19 AM
Kaptive 3 is now integrated within Kaptive-Web (kaptive-web.erc.monash.edu), PathogenWatch (pathogen.watch), the new Kleborate 3 framework (github.com/klebgenomics...) and Bactopia (bactopia.github.io/latest/).
Remember to cite us if you use Kaptive for your results, and watch out for "Untypeable"!
Remember to cite us if you use Kaptive for your results, and watch out for "Untypeable"!
We know the command-line can be tricky, so we made the CLI much friendlier 🧑💻
For the code-savvy, there's also a Python API allowing Kaptive to be used within your own programs 🧱
All the information you need is in the documentation, which we update regularly: kaptive.readthedocs.io/en/latest/
For the code-savvy, there's also a Python API allowing Kaptive to be used within your own programs 🧱
All the information you need is in the documentation, which we update regularly: kaptive.readthedocs.io/en/latest/
Introducing Kaptive 3 — Kaptive 3.0.0 documentation
kaptive.readthedocs.io
February 9, 2025 at 3:19 AM
We know the command-line can be tricky, so we made the CLI much friendlier 🧑💻
For the code-savvy, there's also a Python API allowing Kaptive to be used within your own programs 🧱
All the information you need is in the documentation, which we update regularly: kaptive.readthedocs.io/en/latest/
For the code-savvy, there's also a Python API allowing Kaptive to be used within your own programs 🧱
All the information you need is in the documentation, which we update regularly: kaptive.readthedocs.io/en/latest/
Kaptive 3 is also much (much) faster than Kaptive 2, taking ~1 second per assembly 🏎️💨
This means that if you don't have a fancy HPC, then don't worry! You can still analyse thousands of your own assemblies on your laptop in a reasonable time! 💻
This means that if you don't have a fancy HPC, then don't worry! You can still analyse thousands of your own assemblies on your laptop in a reasonable time! 💻

February 9, 2025 at 3:19 AM
Kaptive 3 is also much (much) faster than Kaptive 2, taking ~1 second per assembly 🏎️💨
This means that if you don't have a fancy HPC, then don't worry! You can still analyse thousands of your own assemblies on your laptop in a reasonable time! 💻
This means that if you don't have a fancy HPC, then don't worry! You can still analyse thousands of your own assemblies on your laptop in a reasonable time! 💻
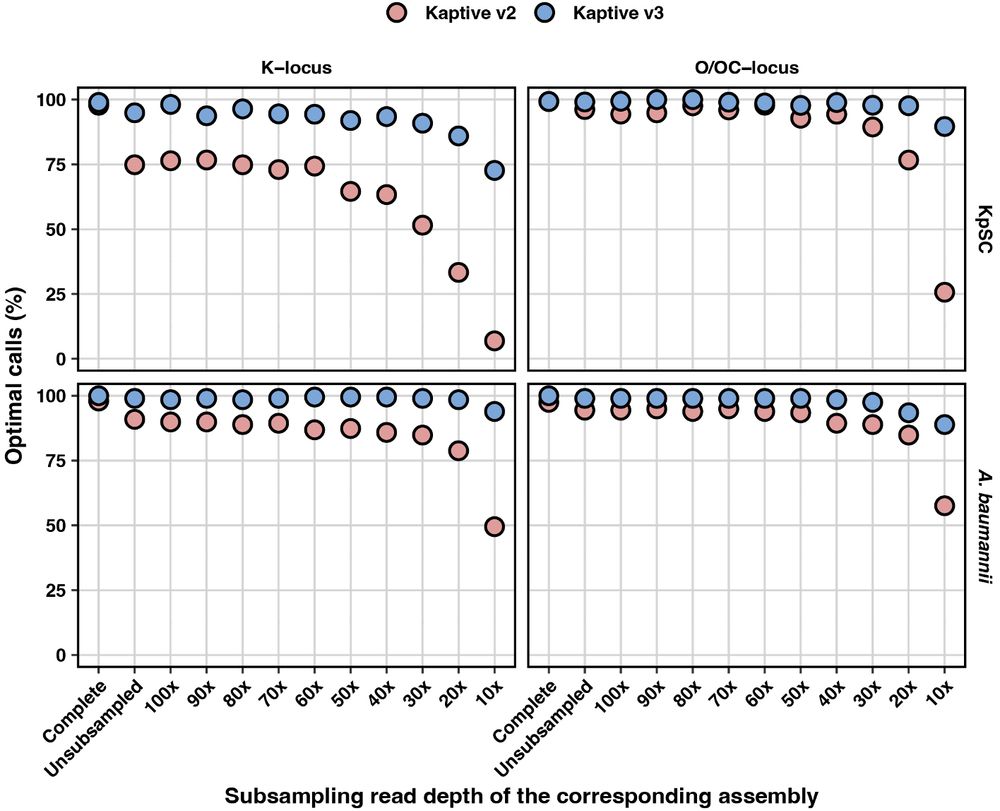
We then subsampled the corresponding short reads at decreasing depths and created sets of increasingly awful draft assemblies with loci broken over contigs and lots of genes missing.
Kaptive 3 was much more sensitive than Kaptive 2, and maintained accuracy even when the assemblies were awful! 💩
Kaptive 3 was much more sensitive than Kaptive 2, and maintained accuracy even when the assemblies were awful! 💩
February 9, 2025 at 3:19 AM
We then subsampled the corresponding short reads at decreasing depths and created sets of increasingly awful draft assemblies with loci broken over contigs and lots of genes missing.
Kaptive 3 was much more sensitive than Kaptive 2, and maintained accuracy even when the assemblies were awful! 💩
Kaptive 3 was much more sensitive than Kaptive 2, and maintained accuracy even when the assemblies were awful! 💩
We put together a special dataset specifically designed to test Kaptive. We had completed hybrid assemblies for KpSC (preprints.scielo.org/index.php/sc...) and A. baumannii (RefSeq).
We identified the K- and O(C)-loci in each and visually confirmed each to determine a ground truth Kaptive call 🔎
We identified the K- and O(C)-loci in each and visually confirmed each to determine a ground truth Kaptive call 🔎
Complete genomes of 568 diverse Klebsiella pneumoniae species complex isolates from humans, animals and marine sources in Norway from 2001-2020
We report 579 hybrid genome assemblies (568 complete) of Klebsiella pneumoniae species complex isolates from human, animal and marine sources in Norway collected 2001-2020, belonging to six phylogroups including K. pneumoniae (n=493) and K. variicola (n=69) and 364 unique sequence types.
preprints.scielo.org
February 9, 2025 at 3:19 AM
We put together a special dataset specifically designed to test Kaptive. We had completed hybrid assemblies for KpSC (preprints.scielo.org/index.php/sc...) and A. baumannii (RefSeq).
We identified the K- and O(C)-loci in each and visually confirmed each to determine a ground truth Kaptive call 🔎
We identified the K- and O(C)-loci in each and visually confirmed each to determine a ground truth Kaptive call 🔎
So enter Kaptive 3, a complete overhaul of Kaptive with a new algorithm designed to handle fragmented loci.
We also refactored (and simplified) the confidence score to be more sensitive for broken loci and missing genes, allowing more Kaptive data to be used when the assembly may not be complete 💯
We also refactored (and simplified) the confidence score to be more sensitive for broken loci and missing genes, allowing more Kaptive data to be used when the assembly may not be complete 💯

February 9, 2025 at 3:19 AM
So enter Kaptive 3, a complete overhaul of Kaptive with a new algorithm designed to handle fragmented loci.
We also refactored (and simplified) the confidence score to be more sensitive for broken loci and missing genes, allowing more Kaptive data to be used when the assembly may not be complete 💯
We also refactored (and simplified) the confidence score to be more sensitive for broken loci and missing genes, allowing more Kaptive data to be used when the assembly may not be complete 💯
Because of how Kaptive 2 chose the best match locus, missing locus sequence resulted in a coverage bias for shorter loci in the database such, and could sometimes lead to inaccurate calls!
Ever seen a stray KL107 in your data that didn't make sense?
Yeah, that's why...
Ever seen a stray KL107 in your data that didn't make sense?
Yeah, that's why...
February 9, 2025 at 3:19 AM
Because of how Kaptive 2 chose the best match locus, missing locus sequence resulted in a coverage bias for shorter loci in the database such, and could sometimes lead to inaccurate calls!
Ever seen a stray KL107 in your data that didn't make sense?
Yeah, that's why...
Ever seen a stray KL107 in your data that didn't make sense?
Yeah, that's why...
So in a nutshell, we traced Kaptive's issues with the Klebsiella K-locus all the way back to the gDNA, where:
The locus region is partially amplified ->
Low sequencing read coverage ->
region doesn't assemble well ->
Untypeable Kaptive call ->
Unusable data 🙅♀️
The locus region is partially amplified ->
Low sequencing read coverage ->
region doesn't assemble well ->
Untypeable Kaptive call ->
Unusable data 🙅♀️
February 9, 2025 at 3:19 AM
So in a nutshell, we traced Kaptive's issues with the Klebsiella K-locus all the way back to the gDNA, where:
The locus region is partially amplified ->
Low sequencing read coverage ->
region doesn't assemble well ->
Untypeable Kaptive call ->
Unusable data 🙅♀️
The locus region is partially amplified ->
Low sequencing read coverage ->
region doesn't assemble well ->
Untypeable Kaptive call ->
Unusable data 🙅♀️
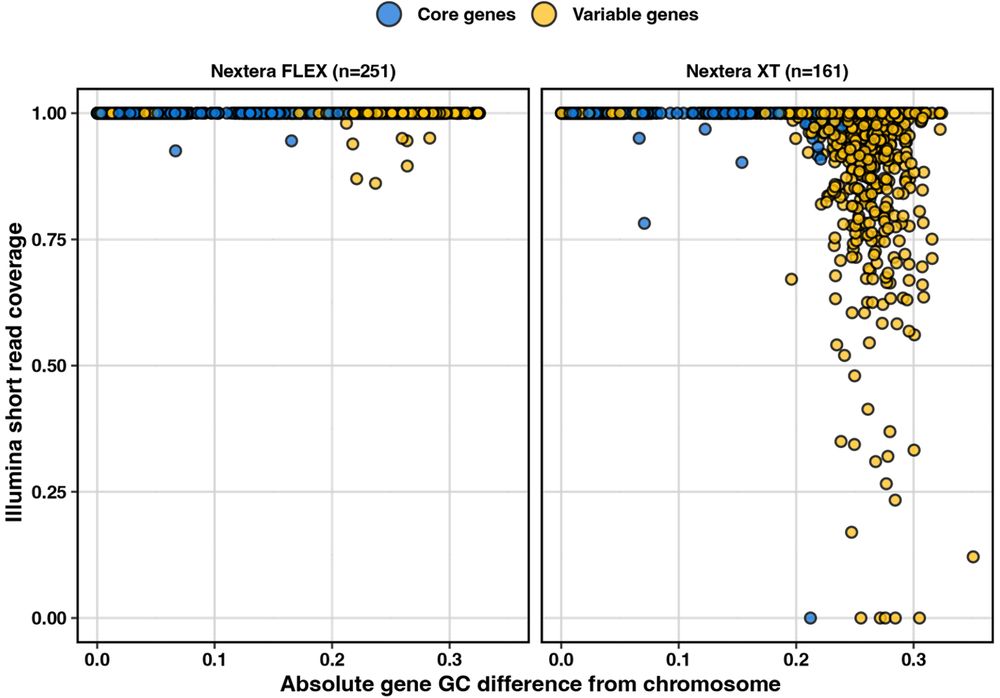
These genes have a very low GC compared to the rest of the Klebsiella chromosome, so we wondered if this was affecting how this part of the genome gets sequenced.
Turns out, these genes show decreased sequencing coverage when reads are prepped with Nextera XT, but not so much with Nextera Flex 🤯
Turns out, these genes show decreased sequencing coverage when reads are prepped with Nextera XT, but not so much with Nextera Flex 🤯
February 9, 2025 at 3:19 AM
These genes have a very low GC compared to the rest of the Klebsiella chromosome, so we wondered if this was affecting how this part of the genome gets sequenced.
Turns out, these genes show decreased sequencing coverage when reads are prepped with Nextera XT, but not so much with Nextera Flex 🤯
Turns out, these genes show decreased sequencing coverage when reads are prepped with Nextera XT, but not so much with Nextera Flex 🤯
The major drivers of low confidence were K-loci that were 1) broken over contigs 🚫 and 2) missing genes ➡️➡️, events that were mostly co-occurring
Turns out, the genes missing were usually those important for antigenic diversity, in this case the glycosyltransferases that dictate the CPS 🍬 structure
Turns out, the genes missing were usually those important for antigenic diversity, in this case the glycosyltransferases that dictate the CPS 🍬 structure
February 9, 2025 at 3:19 AM
The major drivers of low confidence were K-loci that were 1) broken over contigs 🚫 and 2) missing genes ➡️➡️, events that were mostly co-occurring
Turns out, the genes missing were usually those important for antigenic diversity, in this case the glycosyltransferases that dictate the CPS 🍬 structure
Turns out, the genes missing were usually those important for antigenic diversity, in this case the glycosyltransferases that dictate the CPS 🍬 structure
So you may have noticed your Klebsiella K-locus results from previous versions of Kaptive (v1-2) having lots of untypeable calls ("Low" + "None" confidence) with draft assemblies; we certainly did!
This meant that lots of useful seroepi data was unusable, so we started by finding out exactly why 🤔
This meant that lots of useful seroepi data was unusable, so we started by finding out exactly why 🤔
February 9, 2025 at 3:19 AM
So you may have noticed your Klebsiella K-locus results from previous versions of Kaptive (v1-2) having lots of untypeable calls ("Low" + "None" confidence) with draft assemblies; we certainly did!
This meant that lots of useful seroepi data was unusable, so we started by finding out exactly why 🤔
This meant that lots of useful seroepi data was unusable, so we started by finding out exactly why 🤔

