
400 in person and 400 online participants from 71 countries...... Wow!

400 in person and 400 online participants from 71 countries...... Wow!

We defined clone-specific core phenotypes- some common to many clones, some rarer e.g. global MDR SLs 258 and 307 could use fructoselysine, but SL147 couldn't

We defined clone-specific core phenotypes- some common to many clones, some rarer e.g. global MDR SLs 258 and 307 could use fructoselysine, but SL147 couldn't
This was structured so that more closely related isolates shared more similar sets of metabolic traits.
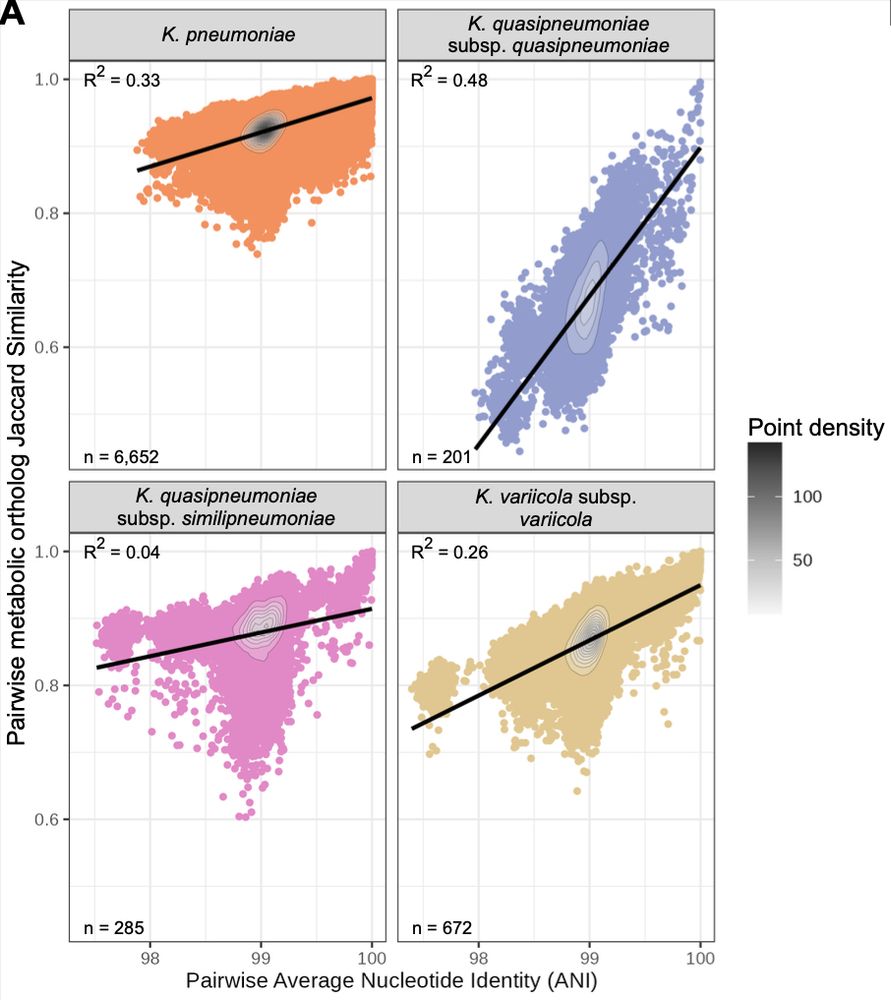
This was structured so that more closely related isolates shared more similar sets of metabolic traits.
Phenotype profiles differed by species - as we expected - and were generally consistent with formal species definitions.

Phenotype profiles differed by species - as we expected - and were generally consistent with formal species definitions.
Most individual traits are either core or very rare - just like genes in the total pan-genome.

Most individual traits are either core or very rare - just like genes in the total pan-genome.


📣📣📣 Congratulations to twitter.com/helena_betha... for preprinting the first chapter of her PhD thesis 🎉🎉🎉
doi.org/10.1101/2023...

📣📣📣 Congratulations to twitter.com/helena_betha... for preprinting the first chapter of her PhD thesis 🎉🎉🎉
doi.org/10.1101/2023...

