www.nature.com/articles/s41...

www.nature.com/articles/s41...

rdcu.be/eKlK7
Led by @dartmouthchem.bsky.social graduate student Kaushilk Borthakur in collaboration with @bonomimax.bsky.social

rdcu.be/eKlK7
Led by @dartmouthchem.bsky.social graduate student Kaushilk Borthakur in collaboration with @bonomimax.bsky.social
The project will focus on molecular modelling of proteins, lipids, and biomolecular condensates at cell membranes.
More details and application form: tinyurl.com/4zm92365
Please feel free to share!
@vetenskapsradet.bsky.social | @mau.se

The project will focus on molecular modelling of proteins, lipids, and biomolecular condensates at cell membranes.
More details and application form: tinyurl.com/4zm92365
Please feel free to share!
@vetenskapsradet.bsky.social | @mau.se
Kinetic NMR screening: rapidly quantifying fast ligand dissociation in fragment mixtures using ¹⁹F relaxation dispersion | doi.org/10.26434/che... 🧲🧪
Kinetic NMR screening: rapidly quantifying fast ligand dissociation in fragment mixtures using ¹⁹F relaxation dispersion | doi.org/10.26434/che... 🧲🧪
doi.org/10.48550/arX...
doi.org/10.48550/arX...

doi.org/10.48550/arX...
A coarse-grained model for simulations of phosphorylated disordered proteins
(aka parameters for phospho-serine and -threonine for CALVADOS)
is now published in Biophysical Journal
authors.elsevier.com/a/1lTcE1SPTB...
@asrauh.bsky.social @giuliotesei.bsky.social & Gustav Hedemark
A coarse-grained model for simulations of phosphorylated disordered proteins
(aka parameters for phospho-serine and -threonine for CALVADOS)
is now published in Biophysical Journal
authors.elsevier.com/a/1lTcE1SPTB...
@asrauh.bsky.social @giuliotesei.bsky.social & Gustav Hedemark
www.biorxiv.org/content/10.1...
Protein Dynamics at Different Timescales Unlock Access to Hidden Post-Translational Modification Sites
#bioinformatics #compchem #folding #proteindynamics

www.biorxiv.org/content/10.1...
Protein Dynamics at Different Timescales Unlock Access to Hidden Post-Translational Modification Sites
#bioinformatics #compchem #folding #proteindynamics


We chatted about grant cancellations, exciting regional meetings and reunions, two fun new preprints, community norms around code release, and the importance of giving kudos. @fraserlab.com
We chatted about grant cancellations, exciting regional meetings and reunions, two fun new preprints, community norms around code release, and the importance of giving kudos. @fraserlab.com
📜 www.biorxiv.org/content/10.1...
🖥️ github.com/KULL-Centre/...

📜 www.biorxiv.org/content/10.1...
🖥️ github.com/KULL-Centre/...
Our design strategy is now published in @natcomms.nature.com
www.nature.com/articles/s41...
#NMRchat #compbio #compchem
Our design strategy is now published in @natcomms.nature.com
www.nature.com/articles/s41...
#NMRchat #compbio #compchem
www.nature.com/articles/s41...
Rational design of fluorine NMR labelling sites (AlphaFold/molecular dynamics) to gain insights into protein structure and interactions for large complexes and in cells!
#NMRchat #compchem #AlphaFold #proteinfolding
www.nature.com/articles/s41...
Rational design of fluorine NMR labelling sites (AlphaFold/molecular dynamics) to gain insights into protein structure and interactions for large complexes and in cells!
#NMRchat #compchem #AlphaFold #proteinfolding
"Characterizing structural and kinetic ensembles of intrinsically disordered proteins using writhe"
www.biorxiv.org/content/10.1...
by Tommy Sisk, with a generative modeling component done in collaboration with @smnlssn.bsky.social
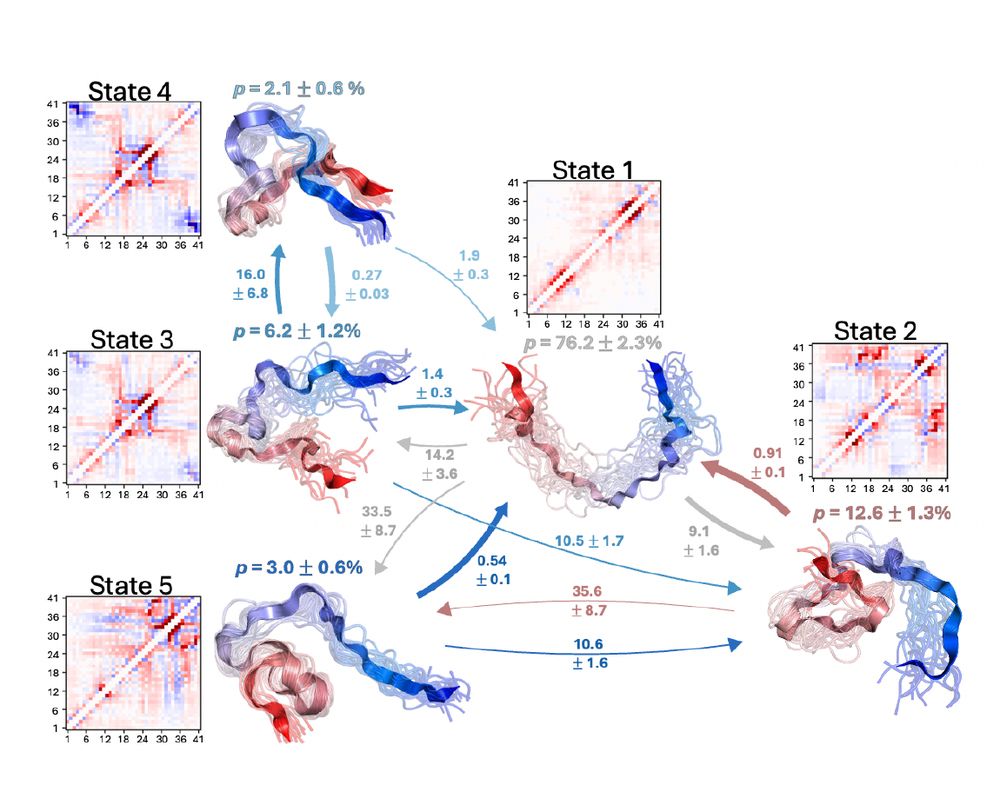
"Characterizing structural and kinetic ensembles of intrinsically disordered proteins using writhe"
www.biorxiv.org/content/10.1...
by Tommy Sisk, with a generative modeling component done in collaboration with @smnlssn.bsky.social
In a fantastic teamwork, @mcagiada.bsky.social and @emilthomasen.bsky.social developed AF2χ to generate conformational ensembles representing side-chain dynamics using AF2 💃
Code: github.com/KULL-Centre/...
Colab: github.com/matteo-cagia...
In a fantastic teamwork, @mcagiada.bsky.social and @emilthomasen.bsky.social developed AF2χ to generate conformational ensembles representing side-chain dynamics using AF2 💃
Code: github.com/KULL-Centre/...
Colab: github.com/matteo-cagia...
www.biorxiv.org/content/10.1...
#proteinfolding #compchem #NMR
www.biorxiv.org/content/10.1...
#proteinfolding #compchem #NMR
Congrats Alki and @twlodarski.bsky.social and co-authors!
Congrats Alki and @twlodarski.bsky.social and co-authors!
www.pnas.org/doi/10.1073/...
The paper has been substantially updated compared to the preprint including new experimental data and using the neural network to finetune CALVADOS. 1/n
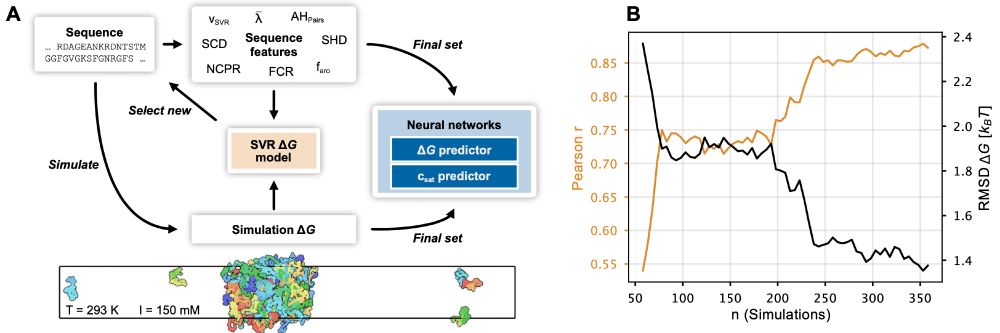
www.pnas.org/doi/10.1073/...
The paper has been substantially updated compared to the preprint including new experimental data and using the neural network to finetune CALVADOS. 1/n
More details about the project including how to apply in the link below. Deadline April 9 2025
candidate.hr-manager.net/ApplicationI...
More details about the project including how to apply in the link below. Deadline April 9 2025
candidate.hr-manager.net/ApplicationI...
@asrauh.bsky.social @giuliotesei.bsky.social and Gustav Hedemark used a top-down approach in which we targeted experimental data to derive parameters or phosphorylated serine and threonine doi.org/10.1101/2025...

@asrauh.bsky.social @giuliotesei.bsky.social and Gustav Hedemark used a top-down approach in which we targeted experimental data to derive parameters or phosphorylated serine and threonine doi.org/10.1101/2025...
#nmrchat #compchem #compbio
doi.org/10.1101/2025.02.10.637539

#nmrchat #compchem #compbio
doi.org/10.1101/2025.02.10.637539


