tinyurl.com/setd2
Special shoutout to @xiaoliwu.bsky.social, jonmarkert.bsky.social, and @ara-latifkar.bsky.social for helping coordinate the event!

Special shoutout to @xiaoliwu.bsky.social, jonmarkert.bsky.social, and @ara-latifkar.bsky.social for helping coordinate the event!


🧬🎉Thrilled to share our new chromatin remodeling study! We reveal three states of human CHD1 and identify a novel "anchor element" that interacts with the acidic patch—conserved among remodelers. Our structures clarify mechanisms of remodeler recruitment! Link: authors.elsevier.com/a/1l2ik3vVUP...
🧬🎉Thrilled to share our new chromatin remodeling study! We reveal three states of human CHD1 and identify a novel "anchor element" that interacts with the acidic patch—conserved among remodelers. Our structures clarify mechanisms of remodeler recruitment! Link: authors.elsevier.com/a/1l2ik3vVUP...

doi.org/10.1016/j.jb... #Chromatin #SIRT6 #CryoEM
tinyurl.com/setd2
PS1: Don't forget that US has switched to PDT
PS2: The recurring registration link:
us06web.zoom.us/webinar/regi...

PS1: Don't forget that US has switched to PDT
PS2: The recurring registration link:
us06web.zoom.us/webinar/regi...
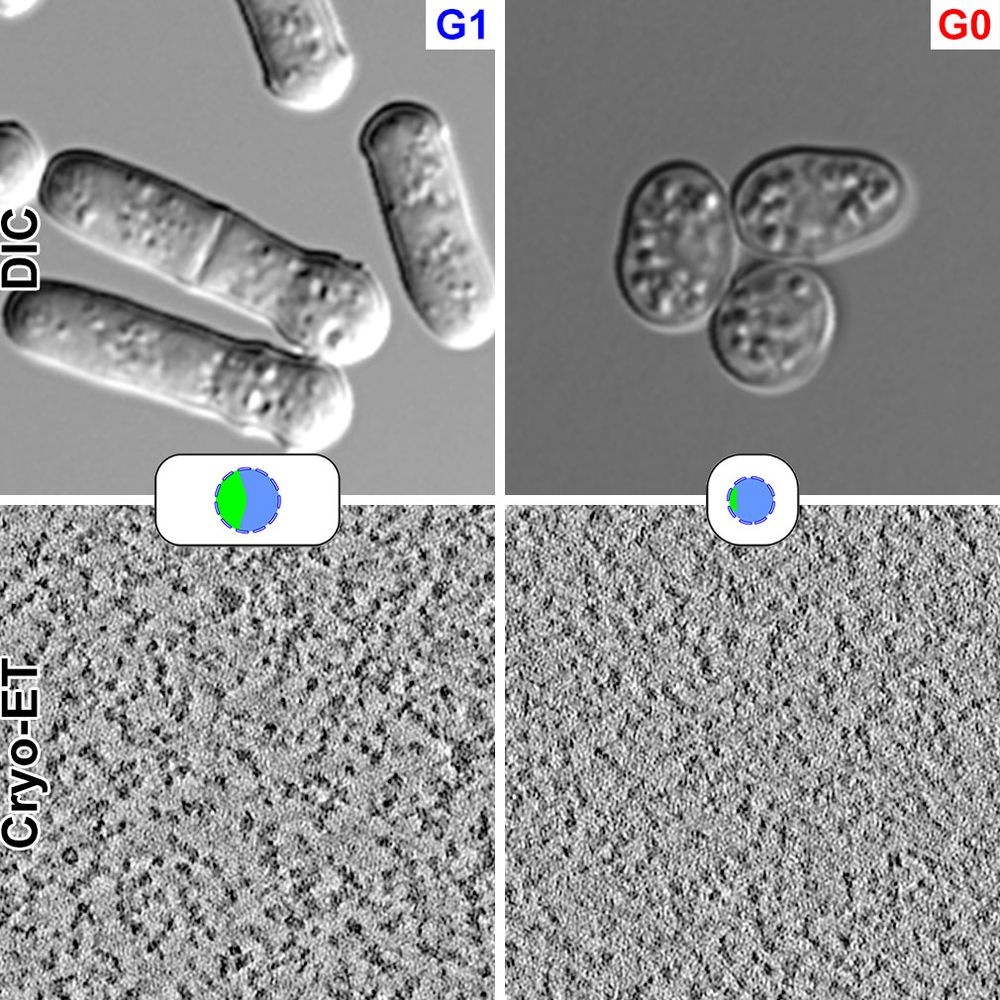
www.science.org/doi/10.1126/...
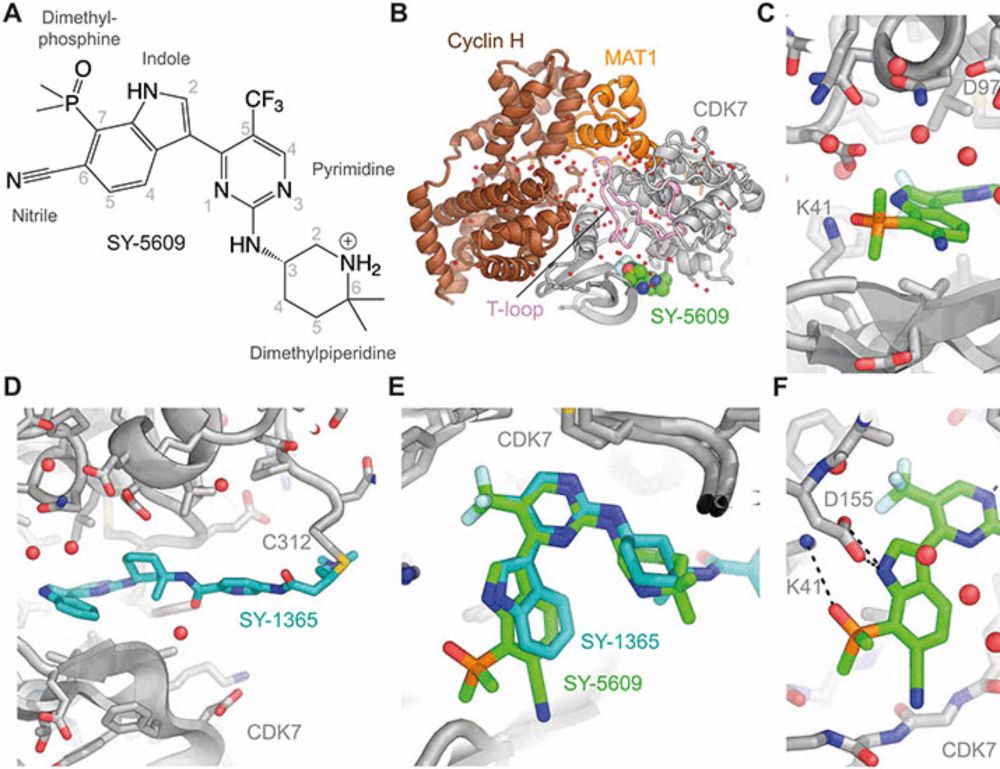
www.science.org/doi/10.1126/...
DM/email me or check farnunglab.com for more info.

DM/email me or check farnunglab.com for more info.
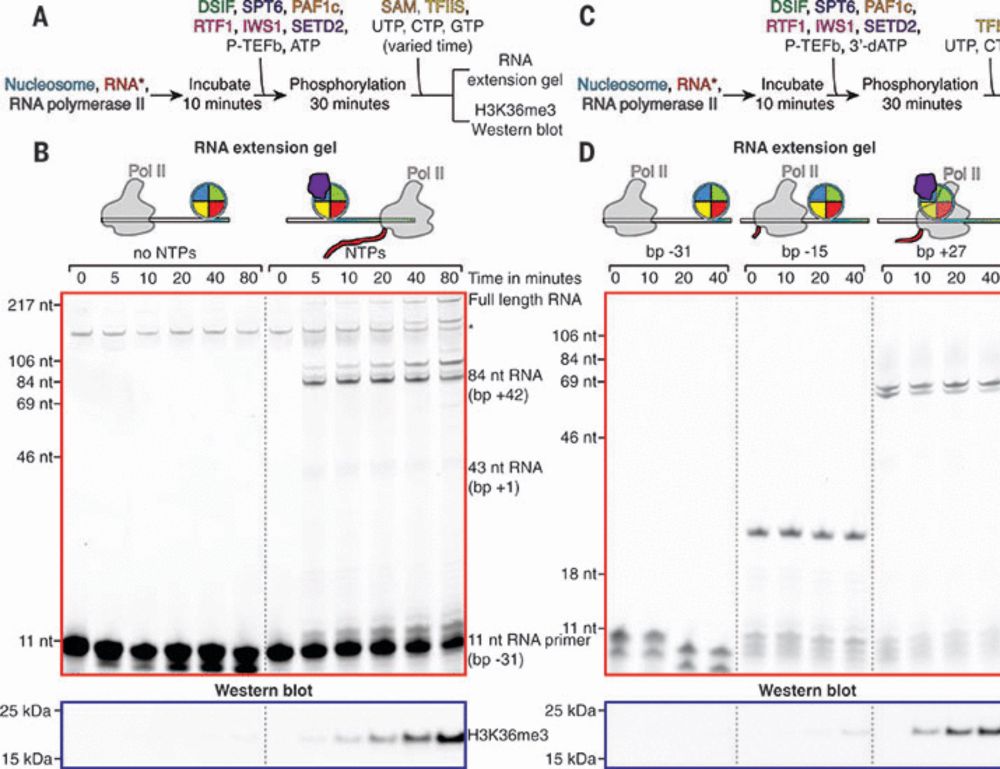




by @jonmarkert.bsky.social @lucas.farnunglab.com et al. at @harvardcellbio.bsky.social #cryoEM
#chromatin #transcription #cryoEM #Pol_II🧪🧬❄️🔬
www.science.org/doi/10.1126/...
by @jonmarkert.bsky.social @lucas.farnunglab.com et al. at @harvardcellbio.bsky.social #cryoEM
#chromatin #transcription #cryoEM #Pol_II🧪🧬❄️🔬
www.science.org/doi/10.1126/...
tinyurl.com/setd2
pubs.acs.org/doi/full/10....
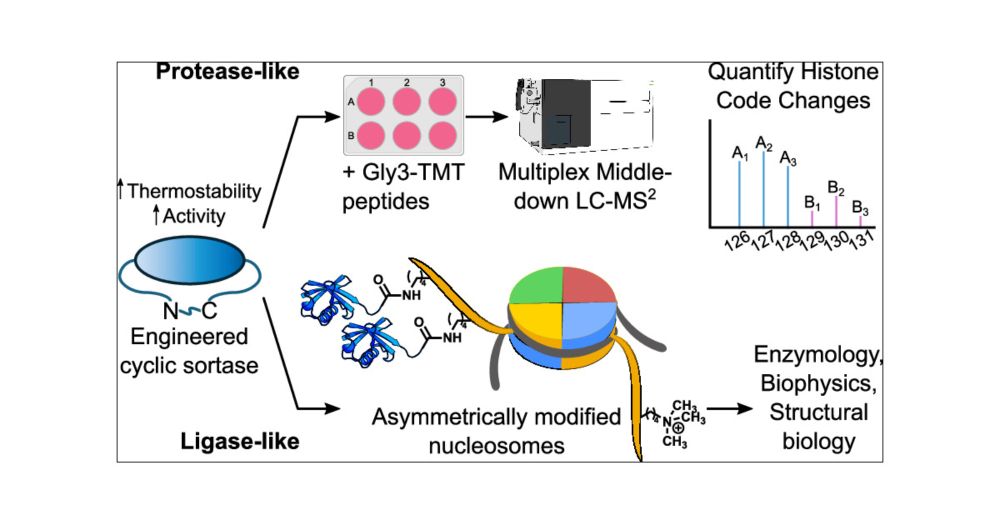
pubs.acs.org/doi/full/10....

