

Go to the paper for cool science, stay for the mesmerizing figures 🤩
Read more about it in this thread 👇
Bentopy makes assembling large-scale MD models accessible and fast.
doi.org/10.1002/pro....
@janstevens.bsky.social, @cg-martini.bsky.social

Go to the paper for cool science, stay for the mesmerizing figures 🤩
Read more about it in this thread 👇
Bentopy makes assembling large-scale MD models accessible and fast.
doi.org/10.1002/pro....
@janstevens.bsky.social, @cg-martini.bsky.social

Bentopy makes assembling large-scale MD models accessible and fast.
doi.org/10.1002/pro....
@janstevens.bsky.social, @cg-martini.bsky.social
If you want to find out how you can save heaps of time while still maintaining accurate results with CG metadynamics, look no further!! Read the article here 👇
If you want to find out how you can save heaps of time while still maintaining accurate results with CG metadynamics, look no further!! Read the article here 👇
Built using bentopy, based on David Naranjo's idea.
The input file can be found here: gist.github.com/ma3ke/723f9c....
Built using bentopy, based on David Naranjo's idea.
The input file can be found here: gist.github.com/ma3ke/723f9c....
qcb.illinois.edu/postdoctoral...
qcb.illinois.edu/postdoctoral...






Consider joining me at the University of Freiburg!
uni-freiburg.de/frias/call-f...
Consider joining me at the University of Freiburg!
uni-freiburg.de/frias/call-f...
www.kearnslab.org


www.kearnslab.org
Upgrading GROMACS to handle billion-atom systems and enhancing I/O performance and precision, making the first-ever whole-cell simulation possible ➡️ bioexcel.eu/uw67
#MolecularDynamics #GROMACS #ComputationalBiology

Upgrading GROMACS to handle billion-atom systems and enhancing I/O performance and precision, making the first-ever whole-cell simulation possible ➡️ bioexcel.eu/uw67
#MolecularDynamics #GROMACS #ComputationalBiology
We studied the opening process of ADAM10 enhanced sampling MD, MSM and Fluorescence microscopy!
Let us know what you think about it 😁
www.biorxiv.org/content/10.1...

We studied the opening process of ADAM10 enhanced sampling MD, MSM and Fluorescence microscopy!
Let us know what you think about it 😁
www.biorxiv.org/content/10.1...
We used molecular dynamics simulations and solid-state NMR (@fmp-berlin.de) to look at calcium binding to the pore domain of the calcium-gated K+ channel MthK. @wojciechkopec.bsky.social @compbiophys.bsky.social
We used molecular dynamics simulations and solid-state NMR (@fmp-berlin.de) to look at calcium binding to the pore domain of the calcium-gated K+ channel MthK. @wojciechkopec.bsky.social @compbiophys.bsky.social

If you like MD simulations, NMR spectroscopy and membranes this story will interest you 😊
"How PGL finds a sweet spot in phospholipid membranes – a combined multiscale MD and NMR study"
www.cell.com/biophysj/abs...

If you like MD simulations, NMR spectroscopy and membranes this story will interest you 😊
📄 Read: pubs.acs.org/doi/10.1021/...
💾 GitHub: github.com/Martini-Forc...
#MolecularDynamics #Biophysics #Simulations #Lipids
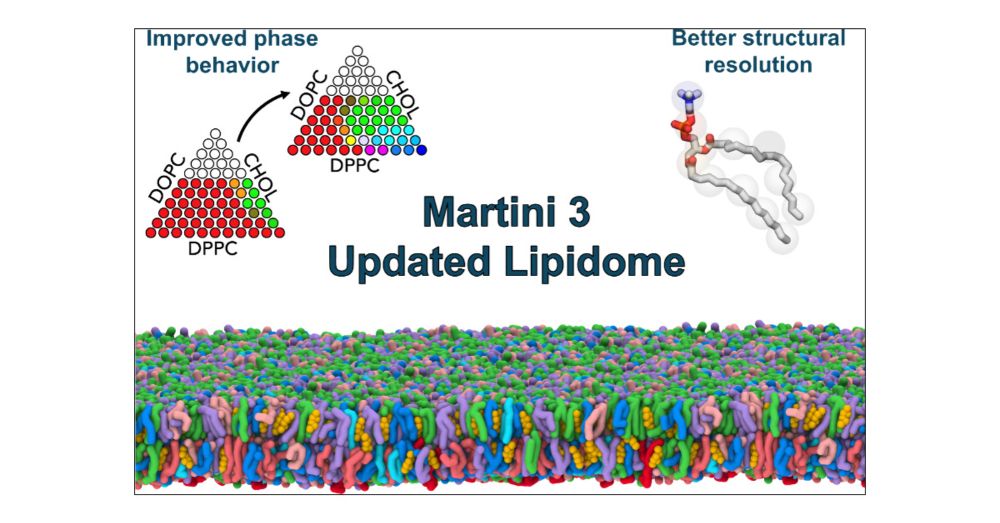
📄 Read: pubs.acs.org/doi/10.1021/...
💾 GitHub: github.com/Martini-Forc...
#MolecularDynamics #Biophysics #Simulations #Lipids
How do nuclear membranes fuse during NPC assembly? We answer this question in our latest work, where we identify a new mechanism for membrane fusion… (1/13)
How do nuclear membranes fuse during NPC assembly? We answer this question in our latest work, where we identify a new mechanism for membrane fusion… (1/13)
Code / Tutorial: github.com/graeter-grou...
Poster: W-109, Thu 17 Jul 11 a.m. PDT — 1:30 p.m. PDT
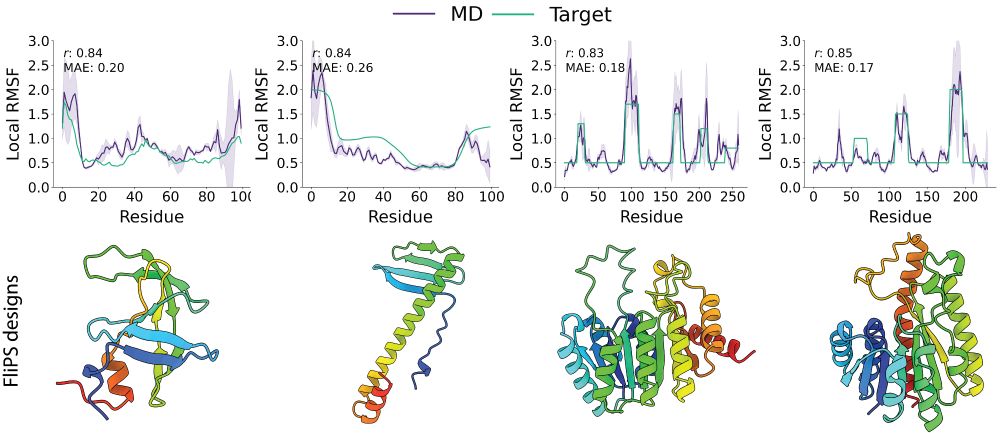
Code / Tutorial: github.com/graeter-grou...
Poster: W-109, Thu 17 Jul 11 a.m. PDT — 1:30 p.m. PDT
Here, we used integrative modelling to build and simulate a mitochondrial cristae.
Find the paper here (rdcu.be/eujAC) or see below for a quick overview 👇
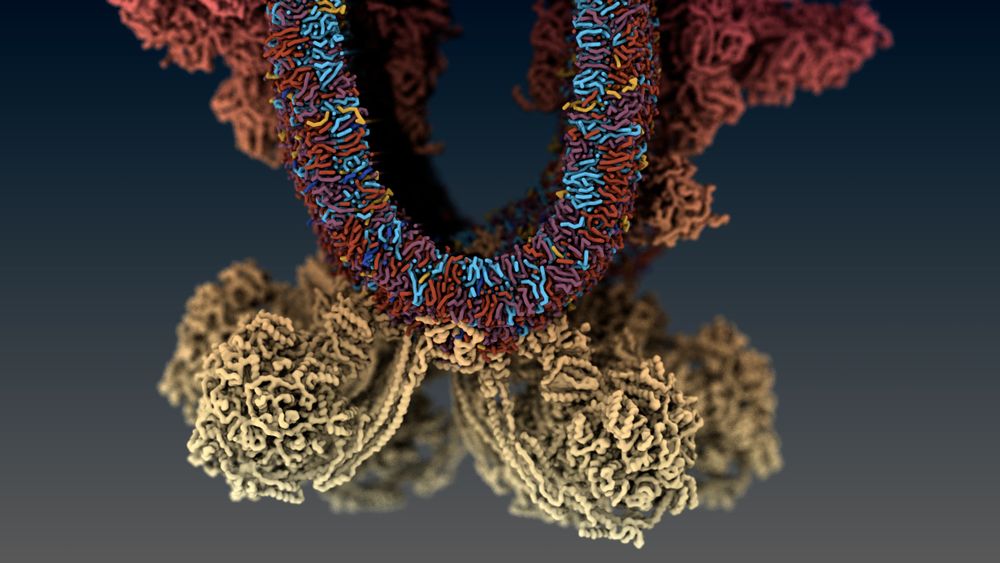
Here, we used integrative modelling to build and simulate a mitochondrial cristae.
Find the paper here (rdcu.be/eujAC) or see below for a quick overview 👇

My passion: proteins, especially enzymes, antibodies, and protein engineering.
Hands-on experience in biochemistry & molecular biology.
Open to exciting opportunities! DMs open 📩
#JobSearch #ProteinEngineering #Postdoc #Biotech
My passion: proteins, especially enzymes, antibodies, and protein engineering.
Hands-on experience in biochemistry & molecular biology.
Open to exciting opportunities! DMs open 📩
#JobSearch #ProteinEngineering #Postdoc #Biotech



