
PhD @GroupParrinello, PostDoc @franknoe.bsky.social
Disordered Proteins, AI for Science, Molecular Dynamics, Enhanced Sampling
🔗 https://scholar.google.com/citations?user=fnJktPAAAAAJ

www.mlsb.io/papers_2024/...
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...

See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
Come help us find new ways to understand and drug intrinsically disordered proteins (IDPs), it's a very interesting and important problem!
Link in the reply ↓
Come help us find new ways to understand and drug intrinsically disordered proteins (IDPs), it's a very interesting and important problem!
Link in the reply ↓
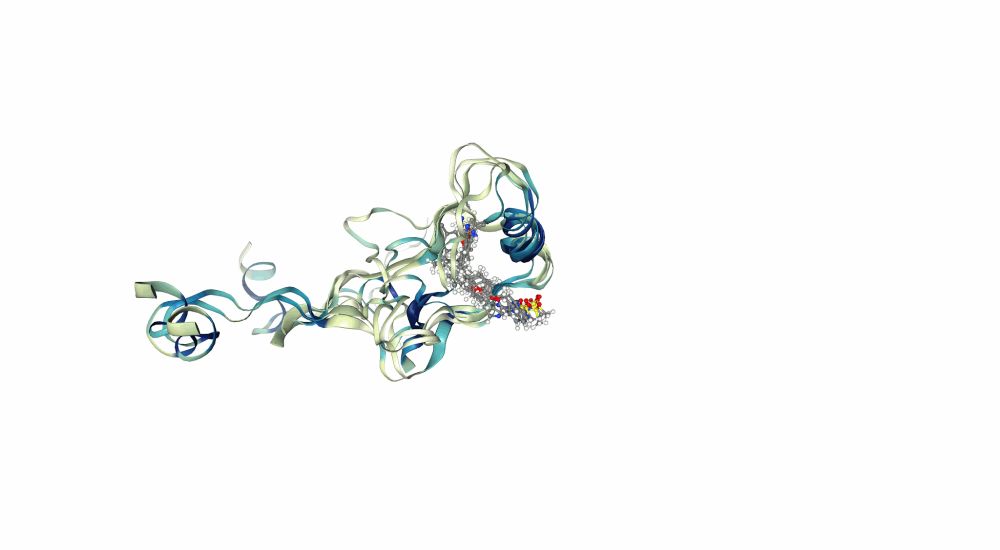
"Characterizing structural and kinetic ensembles of intrinsically disordered proteins using writhe"
www.biorxiv.org/content/10.1...
by Tommy Sisk, with a generative modeling component done in collaboration with @smnlssn.bsky.social
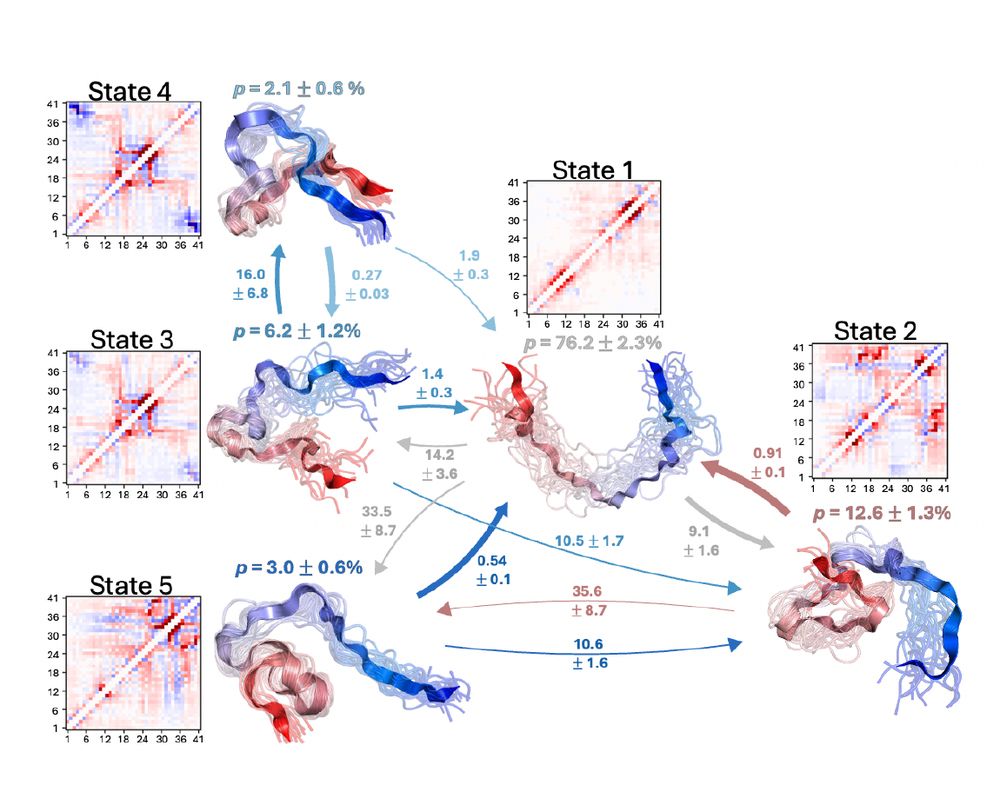
"Characterizing structural and kinetic ensembles of intrinsically disordered proteins using writhe"
www.biorxiv.org/content/10.1...
by Tommy Sisk, with a generative modeling component done in collaboration with @smnlssn.bsky.social
I'll be presenting an overview of the field tomorrow at a workshop. Link to a PDF copy of the presentation: delalamo.xyz/assets/post_...
I'll be presenting an overview of the field tomorrow at a workshop. Link to a PDF copy of the presentation: delalamo.xyz/assets/post_...

We focused here on ligand unbinding kinetics, but this method can be applied to any situation where a reaction coordinate can be defined!

We focused here on ligand unbinding kinetics, but this method can be applied to any situation where a reaction coordinate can be defined!
In a fantastic teamwork, @mcagiada.bsky.social and @emilthomasen.bsky.social developed AF2χ to generate conformational ensembles representing side-chain dynamics using AF2 💃
Code: github.com/KULL-Centre/...
Colab: github.com/matteo-cagia...
In a fantastic teamwork, @mcagiada.bsky.social and @emilthomasen.bsky.social developed AF2χ to generate conformational ensembles representing side-chain dynamics using AF2 💃
Code: github.com/KULL-Centre/...
Colab: github.com/matteo-cagia...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
arxiv.org/html/2503.05...
Useful in particular for de novo designs.
Led by Nico Wolf & Leif Seute, w Seva, Simon, and Jan. @mpip-mainz.mpg.de @hitsters.bsky.social
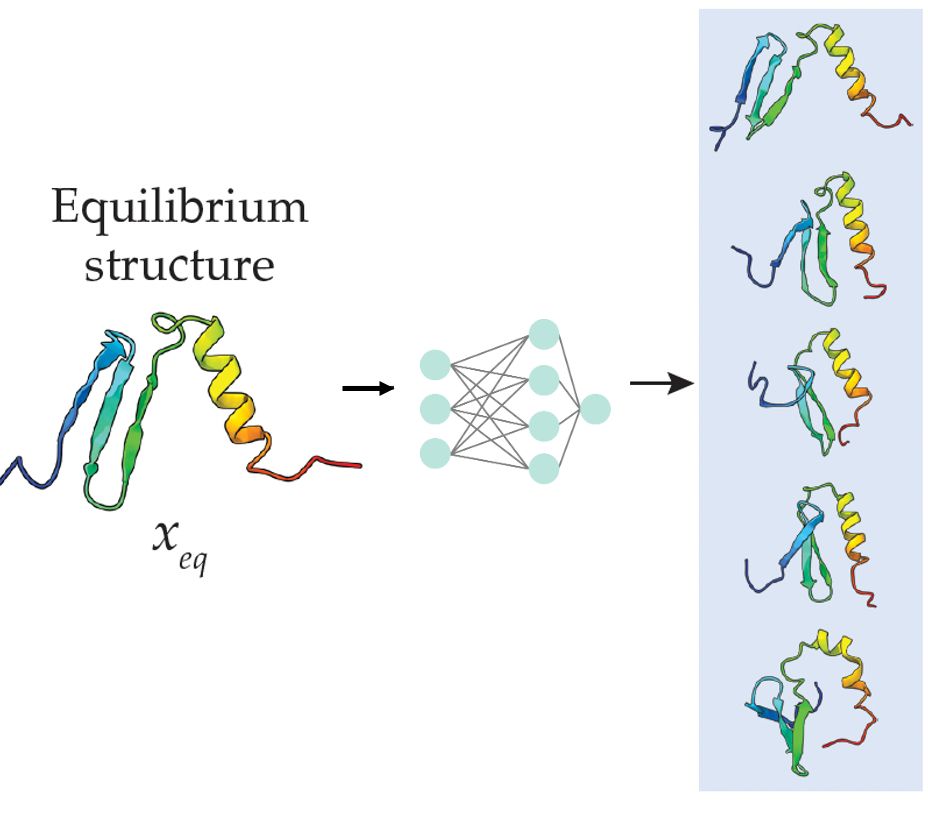
arxiv.org/html/2503.05...
Useful in particular for de novo designs.
Led by Nico Wolf & Leif Seute, w Seva, Simon, and Jan. @mpip-mainz.mpg.de @hitsters.bsky.social
We know proteins fluctuate between different conformations- but by how much? How does it vary from protein to protein? Can highly stable domains have low stability segments? @ajrferrari.bsky.social experimentally tested >5,000 domains to find out!
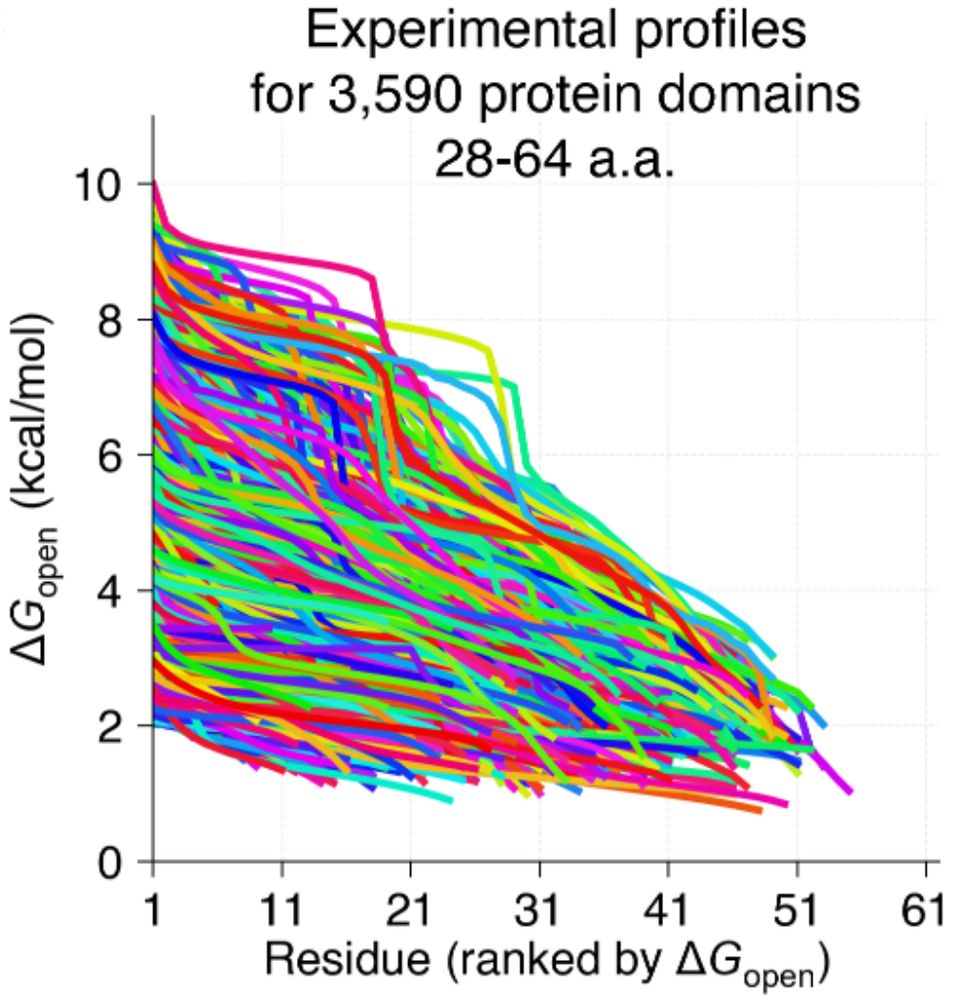
We know proteins fluctuate between different conformations- but by how much? How does it vary from protein to protein? Can highly stable domains have low stability segments? @ajrferrari.bsky.social experimentally tested >5,000 domains to find out!
@hkws.bsky.social and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1!
🧵
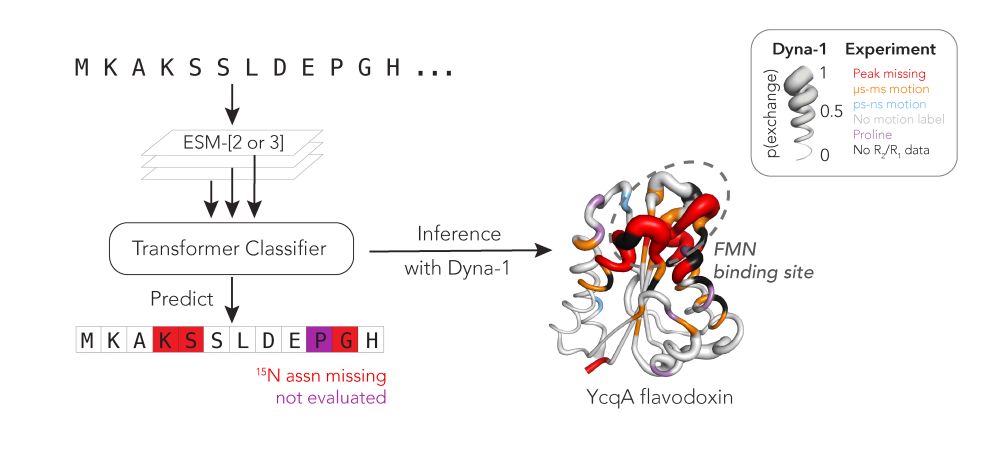
@hkws.bsky.social and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1!
🧵
Training a machine learning model based on residues with missing NMR assignments as a proxy for protein motion
Training a machine learning model based on residues with missing NMR assignments as a proxy for protein motion
Authors: Zhikun Zhang, GiovanniMaria Piccini
DOI: 10.26434/chemrxiv-2025-cvb1v
authors.elsevier.com/sd/article/S...
Led by @sobuelow.bsky.social and Giulio Tesei
authors.elsevier.com/sd/article/S...
Led by @sobuelow.bsky.social and Giulio Tesei

Right now, CADD scientists are forced to use the same model week after week, even if new experimental data says the model is inaccurate.
If we can fine- models, we can exploit that data to systematically improve our predictions week by week!

Right now, CADD scientists are forced to use the same model week after week, even if new experimental data says the model is inaccurate.
If we can fine- models, we can exploit that data to systematically improve our predictions week by week!
Please go ahead, play with it and let us know if there are issues.
github.com/microsoft/bi...
Please go ahead, play with it and let us know if there are issues.
github.com/microsoft/bi...
Mapping Long-Range Interactions in α-Synuclein using Spin-Label NMR and Ensemble Molecular Dynamics Simulations
doi.org/10.1021/ja04...
and I thought I would tell the somewhat random path that led to this paper. 1/n

Mapping Long-Range Interactions in α-Synuclein using Spin-Label NMR and Ensemble Molecular Dynamics Simulations
doi.org/10.1021/ja04...
and I thought I would tell the somewhat random path that led to this paper. 1/n
Could mirror life survive in the wild?
Yes. While mirror life in the wild could have some significant disadvantages (like finding food it can digest), they do not appear to be insurmountable: 🧵

Could mirror life survive in the wild?
Yes. While mirror life in the wild could have some significant disadvantages (like finding food it can digest), they do not appear to be insurmountable: 🧵
“When solving a given problem, try to avoid a more general problem as an intermediate step”
“When solving a given problem, try to avoid a more general problem as an intermediate step”
www.mlsb.io/papers_2024/...

www.mlsb.io/papers_2024/...


