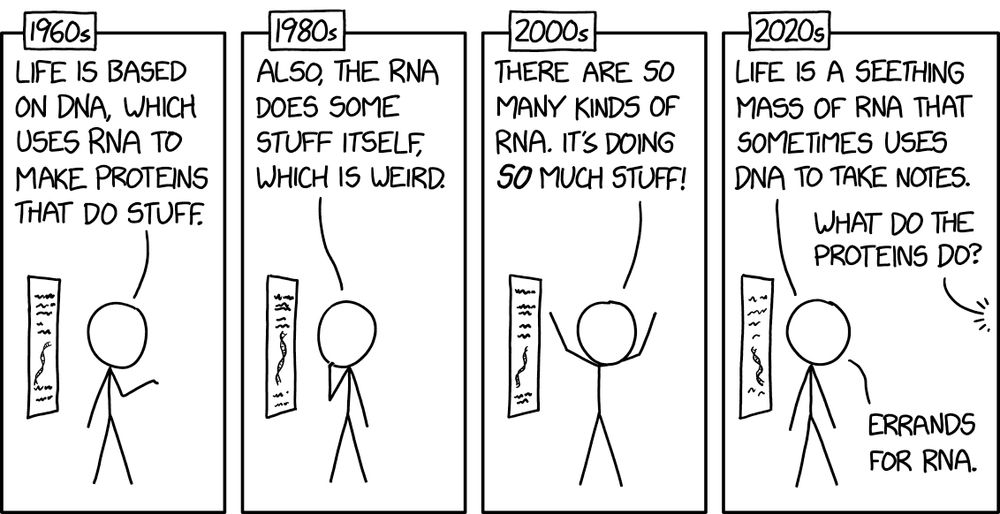
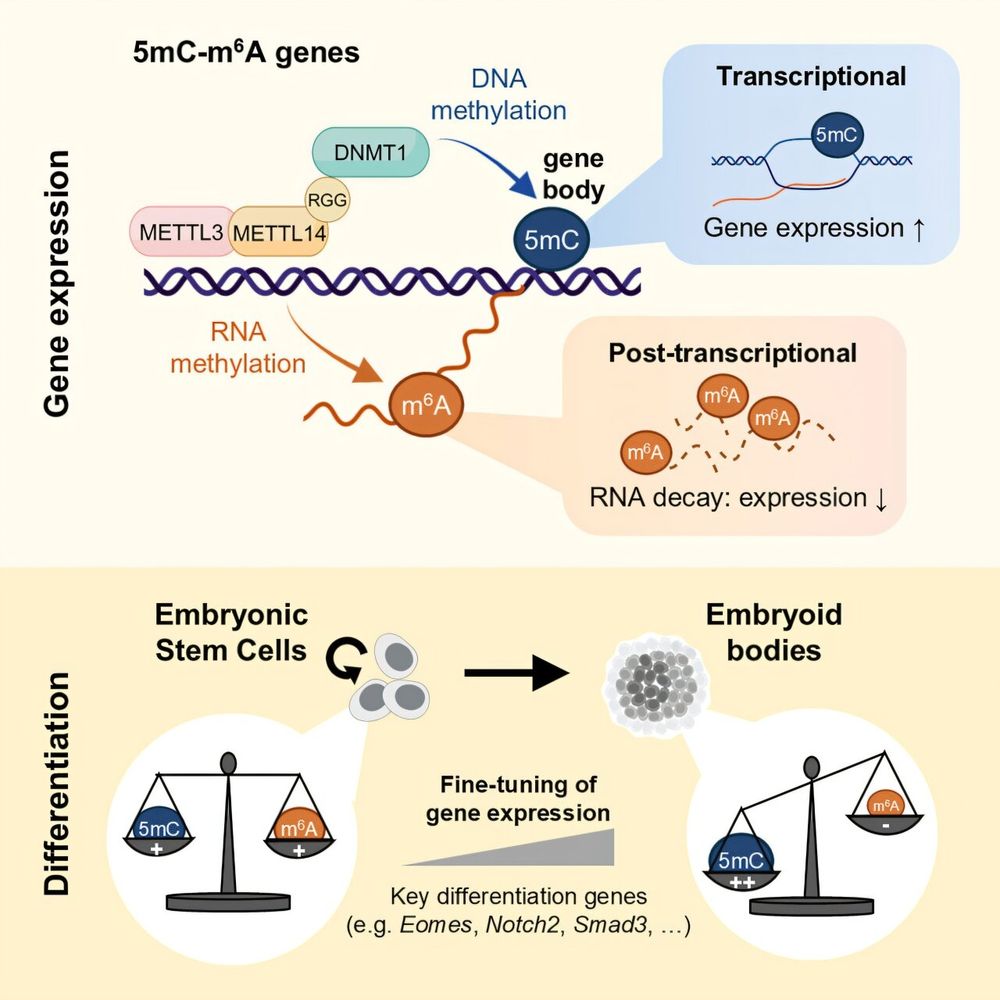
andreagomezlab.com

Check out our searchable gene database and look up your favorite gene 🧬
www.nature.com/articles/s41...

Check out our searchable gene database and look up your favorite gene 🧬
www.nature.com/articles/s41...

I'll put it at the top of my reading list, but the abstract already gave me some cool ideas.
#RNASky #RNABiology 🧪
www.biorxiv.org/content/10.1...
I'll put it at the top of my reading list, but the abstract already gave me some cool ideas.
#RNASky #RNABiology 🧪
www.biorxiv.org/content/10.1...

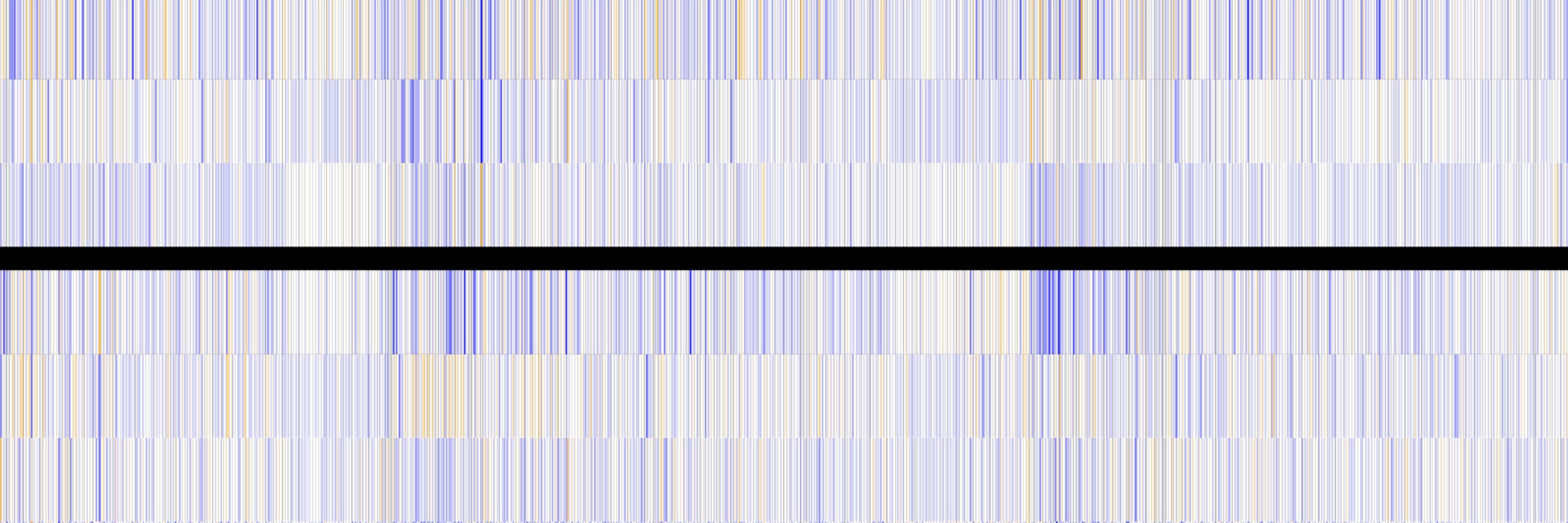
We show that psychedelics induce subtle changes in gene expression but robust changes in alternative splicing lasting at least one month, suggesting that alternative splicing may represent a general mechanism for prolonged plasticity in neurons. #RNAsky
We show that psychedelics induce subtle changes in gene expression but robust changes in alternative splicing lasting at least one month, suggesting that alternative splicing may represent a general mechanism for prolonged plasticity in neurons. #RNAsky