Computational Biologist at LMU München. RevBayes, phylogenetics, Bayes & Macroevolution. Former Miller fellow (UC Berkeley), Emmy Noether group leader & ERC StG
Reposted by Sebastian Höhna

Reposted by Sebastian Höhna
See
job-portal.lmu.de/jobposting/6...
Reposted by Sebastian Höhna
👉 bornberglab.org/post-doc-pos...
Reposted by Sebastian Höhna

by Dr Stephan Spiekman
Tuesday | 25.11.2025 | 16:00
Join the talk in person at the Museum am Löwentor or online: https://t1p.de/sq0m6
Reposted by Sebastian Höhna
Join us at @uni-goettingen.de for a 3-year PhD (65% TV-L E13).
We will investigate the genomic & phenotypic impact of gene duplication across 233 arthropod genomes!
More infos 👇 and s.gwdg.de/eDrAAY
#Evolution #Genomics #Bioinformatics
Reposted by Sebastian Höhna

Reposted by Sebastian Höhna
Reposted by Sebastian Höhna

Find out more: royalsocietypublishing.org/doi/10.1098/... #evolution #palaeontology #taxonomyandsystematics
Reposted by Simon J. Greenhill, Sebastian Höhna

Reposted by Sebastian Höhna, Orlando Schwery

Reposted by Wolfgang Kiessling, Sebastian Höhna

Among our 29 themed symposia + 1 open symposium, we are pleased to feature:
✨ Life in the Phanerozoic Oceans: Evolution, diversity and ecology in deep time marine ecosystems✨
📩 To participate in this symposium or get more information, contact the conveners: 👇
Reposted by Sebastian Höhna

(I would still love to see a densitree plot though!)
www.biorxiv.org/content/10.1...
Reposted by Sebastian Höhna




So incredible to watch our undergrad students from the lab shine in front of such a knowledgeable audience at @cpeg-cpb25.bsky.social. They absolutely crushed it and I couldn’t be prouder!
@fernandalandim.bsky.social, Anna Clara Annes, Jorge Silva and Gabriela Karam.
Reposted by Sebastian Höhna

We are putting together an amazing workshop with details to come - can't wait to share it with everyone!🥳
Reposted by Sebastian Höhna

Reposted by John R. Hutchinson, Sebastian Höhna

www.nature.com/articles/d41...
Reposted by Sebastian Höhna

Reposted by Christian Hof, Sebastian Höhna

Ad here: jobs.uni-rostock.de/jobposting/c...
Reposted by Sebastian Höhna

go.nature.com/4lFx4KN
Reposted by Sebastian Höhna

We introduce a covarion model for phylogenetic inference using discrete morphological data, addressing lineage- and character-specific rate heterogeneity.
www.biorxiv.org/content/10.1...
#phylogenetics #morphology #covarion #evolution
Reposted by Sebastian Höhna
Reposted by Sebastian Höhna
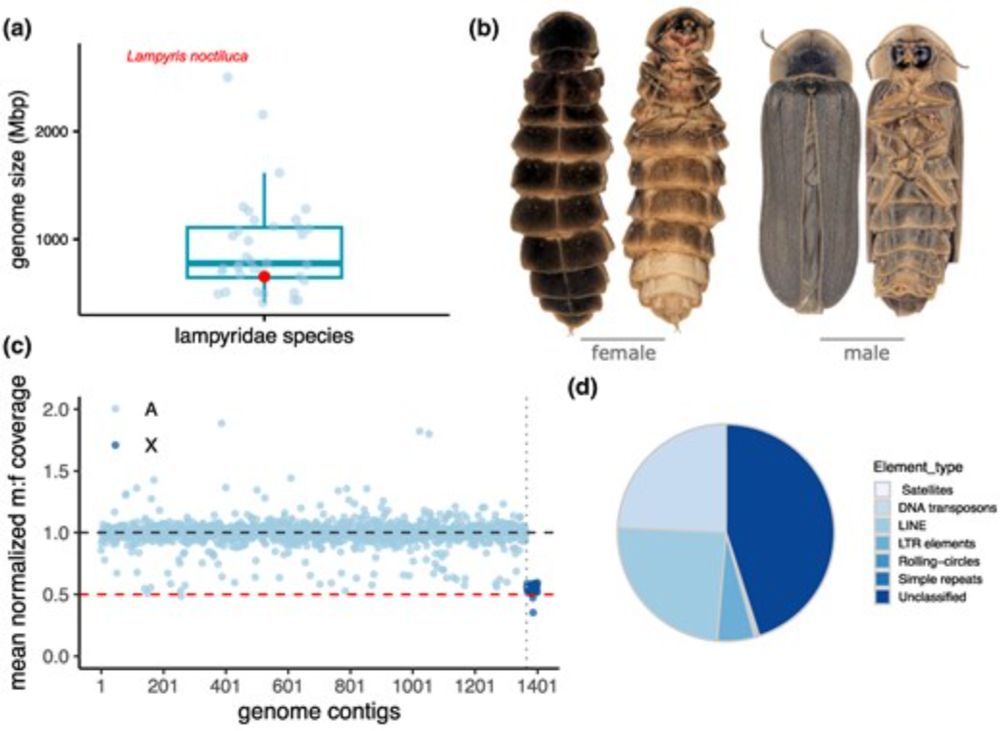
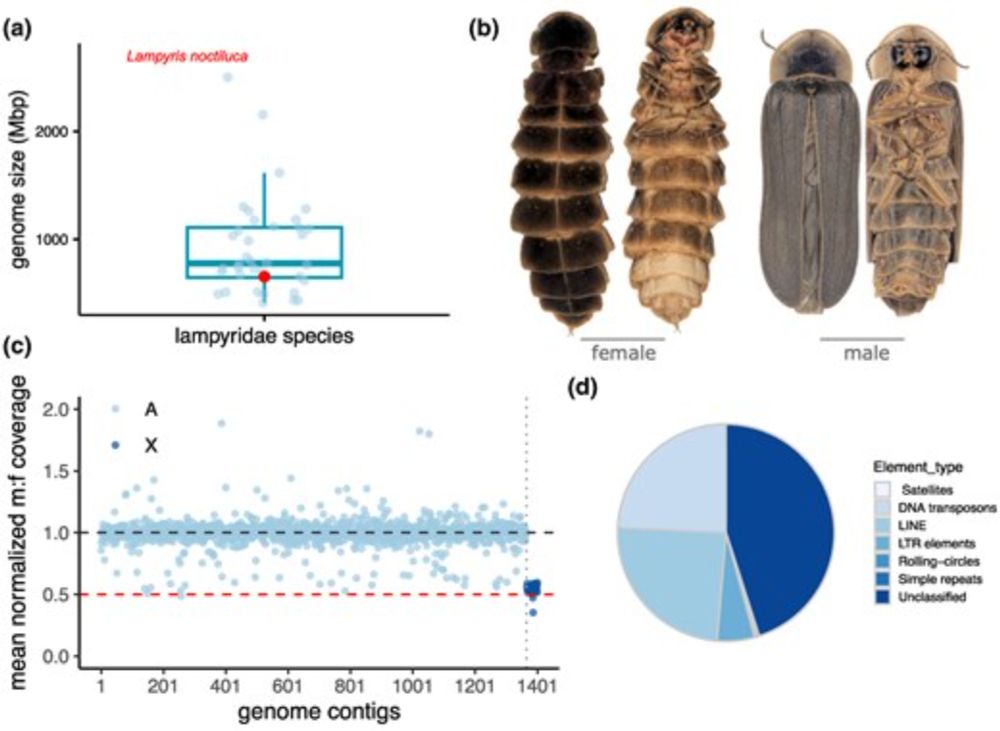
🔗 academic.oup.com/mbe/article/42/6/msaf123/8170119
#evobio #molbio
Reposted by Sebastian Höhna

Model. Preprint coming soon!
Reposted by Sebastian Höhna

Reposted by Sebastian Höhna
📍 Museum für Naturkunde, Berlin, Germany
🕒 3 Years, fully funded by the Leibniz Junior Researchgroup program
Details and Application portal 👇
jobs.museumfuernaturkunde.berlin/jobposting/8...

