

🌴 🐒
Paper: arxiv.org/abs/2501.05457
Code: github.com/molML/jungle...
#ChemBio #ChemSky

#ChemBio #ChemSky
Organizers:
🔸 Nadine Schneider
🔸 @fragrisoni.bsky.social
🔸 José Miguel Hernández Lobato
Organizers:
🔸 Nadine Schneider
🔸 @fragrisoni.bsky.social
🔸 José Miguel Hernández Lobato
Our speakers are truly passionate about driving towards a positive change! 🌍
Special congratulations to @fragrisoni.bsky.social, the very first recipient of the Green Dream Reactions Award!! 🏆👏
#DreamReactions#Sustainability

Our speakers are truly passionate about driving towards a positive change! 🌍
Special congratulations to @fragrisoni.bsky.social, the very first recipient of the Green Dream Reactions Award!! 🏆👏
#DreamReactions#Sustainability


@machine.learning.bio Christian Dallago, Duke University, US
@delafuentelab.bsky.social Cesar de la Fuente, UPenn, US
@fragrisoni.bsky.social Francesca Grisoni - TU/e, NL
Birte Hoecker, Bayeruth University, DE
@machine.learning.bio Christian Dallago, Duke University, US
@delafuentelab.bsky.social Cesar de la Fuente, UPenn, US
@fragrisoni.bsky.social Francesca Grisoni - TU/e, NL
Birte Hoecker, Bayeruth University, DE
Find it here: https://doi.org/10.1093/bioadv/vbaf058
Authors include: @fragrisoni.bsky.social
Find it here: https://doi.org/10.1093/bioadv/vbaf058
Authors include: @fragrisoni.bsky.social
Using joint modeling, we could detect distribution shifts, estimate prediction reliability, and capture meaningful molecular patterns!
doi.org/10.26434/che...
#AI #chemistry

Using joint modeling, we could detect distribution shifts, estimate prediction reliability, and capture meaningful molecular patterns!
doi.org/10.26434/che...
#AI #chemistry
There is an ever growing list of amazing speakers confirmed for this event. (1/2)
#chemsky #ChemBio @chemistryeurope.bsky.social
www.cbc-cpc2025.org
There is an ever growing list of amazing speakers confirmed for this event. (1/2)
#chemsky #ChemBio @chemistryeurope.bsky.social
www.cbc-cpc2025.org

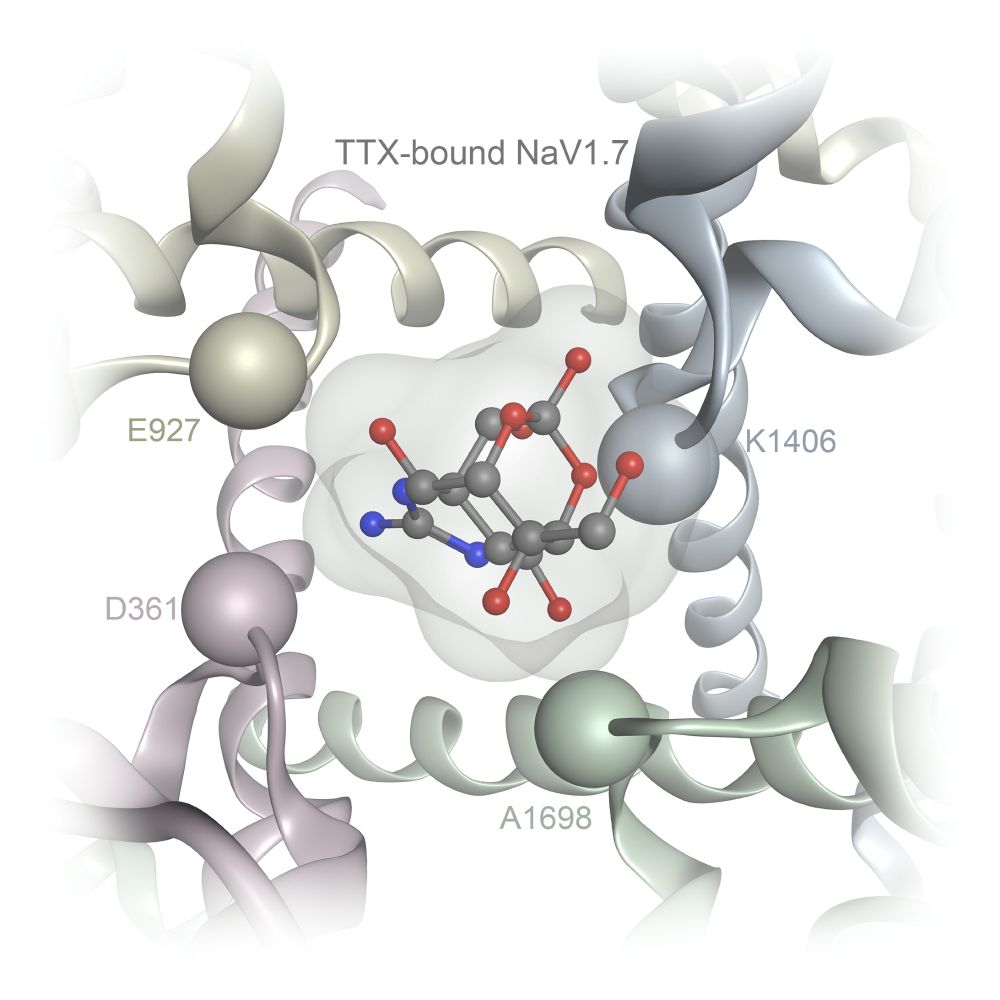
jcheminf.biomedcentral.com/articles/10....
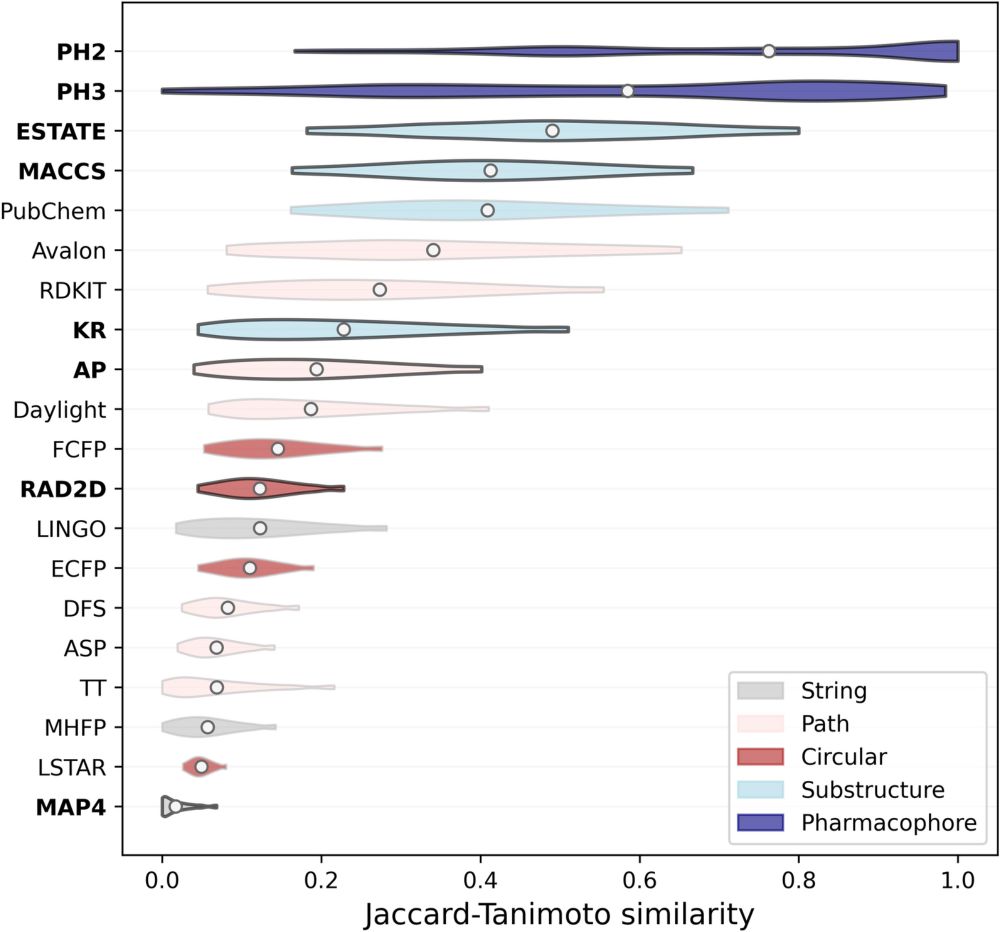
jcheminf.biomedcentral.com/articles/10....

🌴 🐒
Paper: arxiv.org/abs/2501.05457
Code: github.com/molML/jungle...

🌴 🐒
Paper: arxiv.org/abs/2501.05457
Code: github.com/molML/jungle...
Funded by the European Research Council (ERC).
Deadline: Jan 15, 25
jobs.tue.nl/nl/vacature/...
Repost appreciated!

Funded by the European Research Council (ERC).
Deadline: Jan 15, 25
jobs.tue.nl/nl/vacature/...
Repost appreciated!
Authors: Emanuele Criscuolo, Rıza Özçelik, Derek van Tilborg, Francesca Grisoni
DOI: 10.26434/chemrxiv-2024-rp81v
Authors: Emanuele Criscuolo, Rıza Özçelik, Derek van Tilborg, Francesca Grisoni
DOI: 10.26434/chemrxiv-2024-rp81v
Authors: Yannick Leurs, Willem van den Hout, Andrea Gardin, Joost van Dongen, Jan van Hest, Francesca Grisoni, Luc Brunsveld
DOI: 10.26434/chemrxiv-2024-frnj3
Authors: Yannick Leurs, Willem van den Hout, Andrea Gardin, Joost van Dongen, Jan van Hest, Francesca Grisoni, Luc Brunsveld
DOI: 10.26434/chemrxiv-2024-frnj3
In "Learning on compressed molecular representations" Jan Weinreich and I looked into whether GZIP performed better than Neural Networks in chemical machine learning tasks. Yes, you've read that right.
TL;DR: Yes, GZIP can perform better than baseline GNNs and MLPs. It can ..

In "Learning on compressed molecular representations" Jan Weinreich and I looked into whether GZIP performed better than Neural Networks in chemical machine learning tasks. Yes, you've read that right.
TL;DR: Yes, GZIP can perform better than baseline GNNs and MLPs. It can ..

