
Preprint: www.biorxiv.org/content/10.1...
(1/10)
Preprint: www.biorxiv.org/content/10.1...
(1/10)

Preprint: www.biorxiv.org/content/10.1...
(1/10)
TRACeR for MHC-I: go.nature.com/4gcLzn5
TRACeR for MHC-II: go.nature.com/4gj5OQk
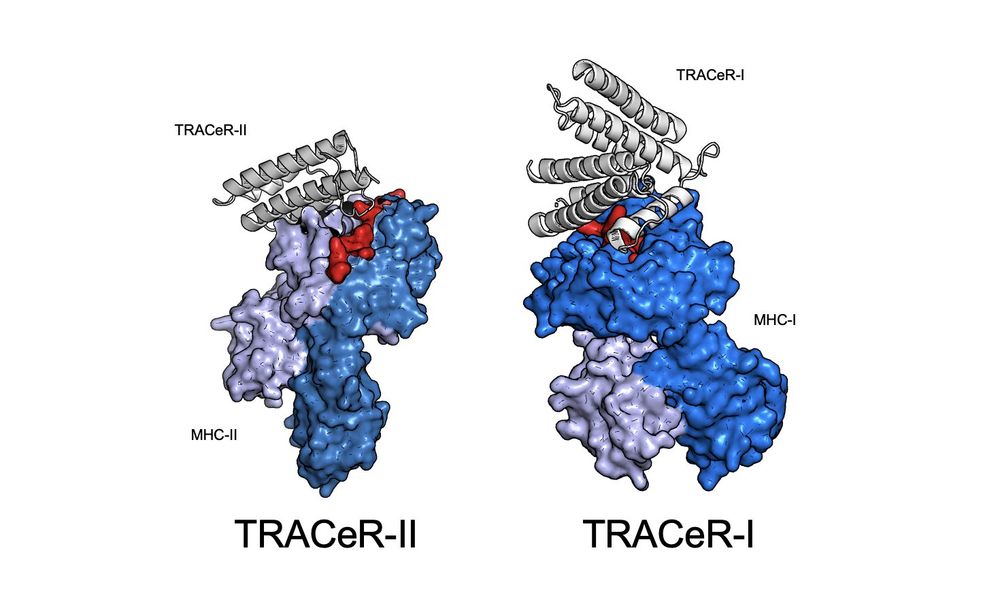
TRACeR for MHC-I: go.nature.com/4gcLzn5
TRACeR for MHC-II: go.nature.com/4gj5OQk
DMSO= helps but kills pol processivity
Betaine= Lots of spurious products
1,2-propandiol= processivity retained and usually specific product
Gel order:
Upstream homology arm, downstream homology arm, full length locus with target gene in middle, a not so great detection primer set. Ladders are NEB 1KB Plus. Arms 1kbp. Full locus 2880bp. All amplified in same conditions.
10/n




DMSO= helps but kills pol processivity
Betaine= Lots of spurious products
1,2-propandiol= processivity retained and usually specific product
#proteomics #bioinformatics
#proteomics #bioinformatics
blueskydirectory.com/starter-pack...

blueskydirectory.com/starter-pack...
#MolecularBiology

#MolecularBiology
Complete with collective nouns
I’ve been tracking these but now can’t keep up
Hope it’s helpful
2/2
docs.google.com/spreadsheets...
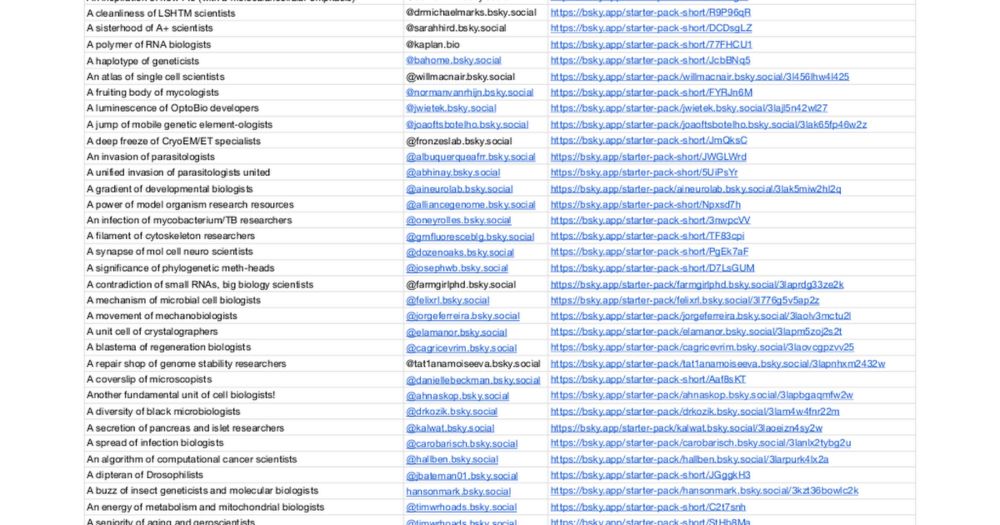
Complete with collective nouns
I’ve been tracking these but now can’t keep up
Hope it’s helpful
2/2
docs.google.com/spreadsheets...

