www.nature.com/articles/s41...
#AI #ProteinDesign #AIPDP




#AI #ProteinDesign #AIPDP

Our new preprint identifies Elephant Herpesvirus gH/gL/gO as a receptor-binding complex and a promising vaccine candidate, now being tested in young elephants across European zoos! 💉
www.biorxiv.org/content/10.6...
Our new preprint identifies Elephant Herpesvirus gH/gL/gO as a receptor-binding complex and a promising vaccine candidate, now being tested in young elephants across European zoos! 💉
www.biorxiv.org/content/10.6...
We found that BAM in Bacteroides and Porphyromonas gingivalis has a distinct architecture from BAM in Proteobacteria.
doi.org/10.1038/s415...

We found that BAM in Bacteroides and Porphyromonas gingivalis has a distinct architecture from BAM in Proteobacteria.
doi.org/10.1038/s415...
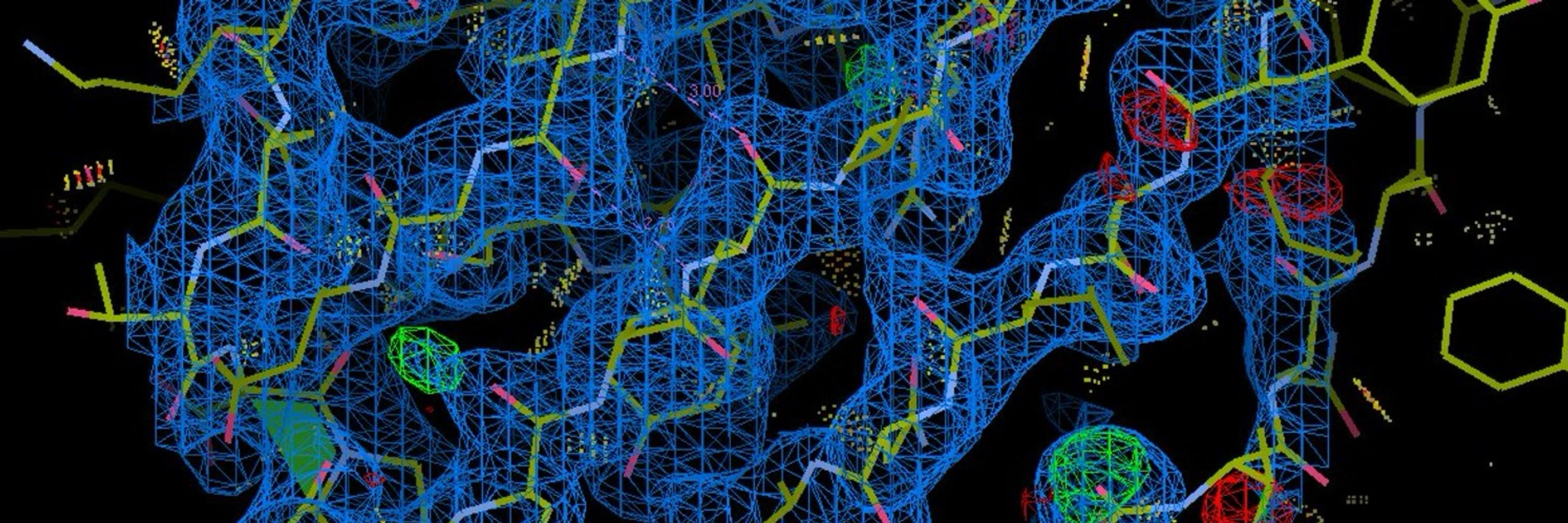
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
www.cell.com/structure/fu...

www.cell.com/structure/fu...
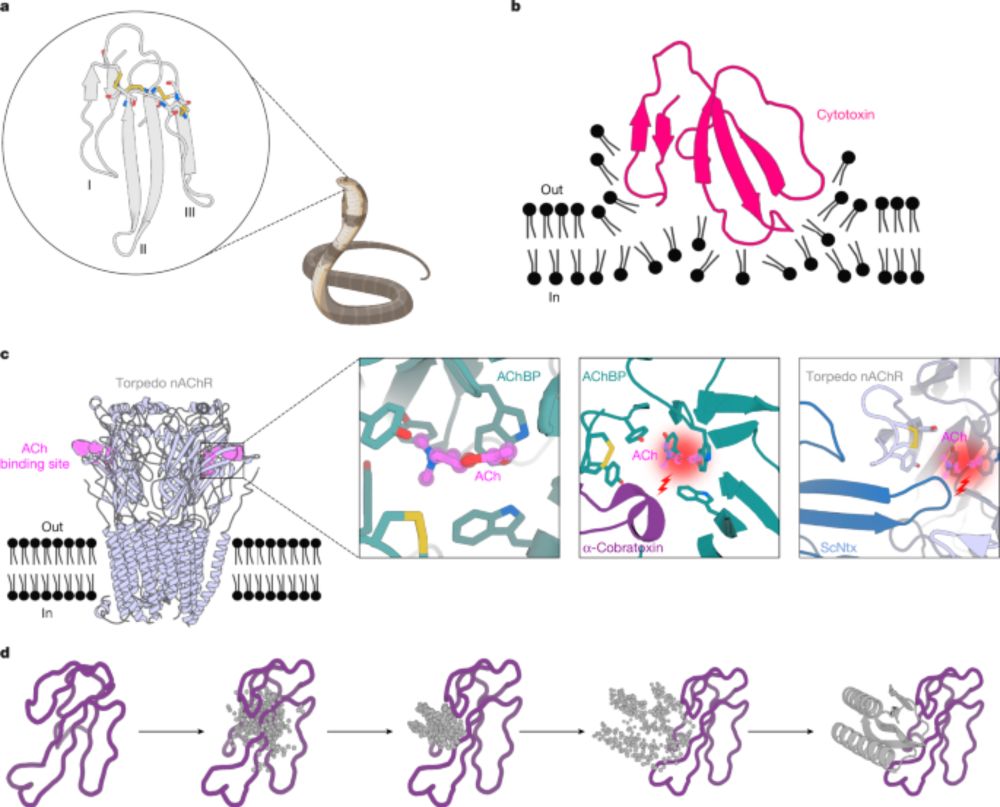
It covers the history of this work + has lots of good case studies, including how these tools are being used to make snake anti-venoms.
The tables are particularly valuable.




It covers the history of this work + has lots of good case studies, including how these tools are being used to make snake anti-venoms.
The tables are particularly valuable.

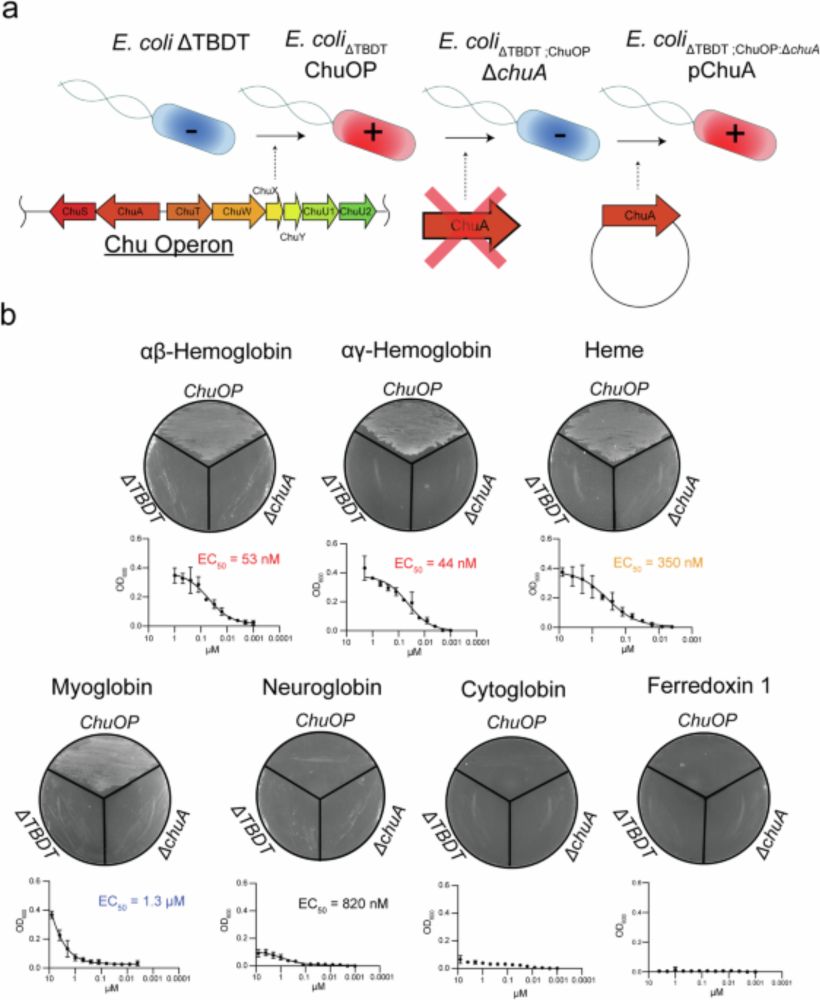
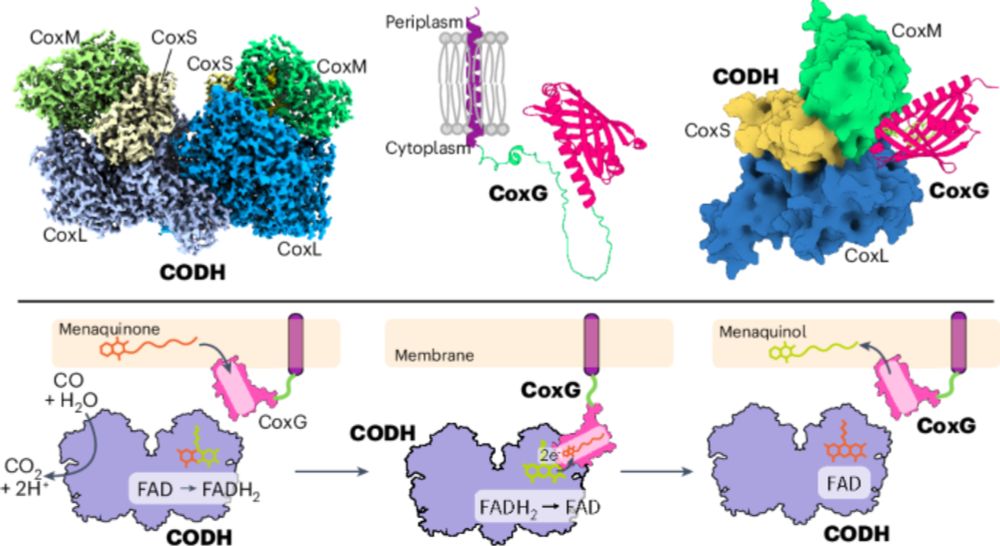
In PNAS today, we show how bacteria can do this using just three enzymes. Honoured to contribute to this study with Christoph von Ballmoos, Chris Greening and Gregory Cook
lnkd.in/gxm8SXxF
In PNAS today, we show how bacteria can do this using just three enzymes. Honoured to contribute to this study with Christoph von Ballmoos, Chris Greening and Gregory Cook
lnkd.in/gxm8SXxF
Preprint 📝: biorxiv.org/content/10.1...
🧵 1/n
Preprint 📝: biorxiv.org/content/10.1...
🧵 1/n
You can sign up for future event information and reminders here:
australian-structural-biology-computing.github.io/website/
🧶🧬

You can sign up for future event information and reminders here:
australian-structural-biology-computing.github.io/website/
🧶🧬
www.nature.com/articles/s41...

www.nature.com/articles/s41...


It was a great opportunity to network with other students and ECRs and was completely inspired by all the cool science🔬🧬




It was a great opportunity to network with other students and ECRs and was completely inspired by all the cool science🔬🧬
We’re losing species we love & major parties are sleepwalking through it.
Australians care. It’s time politicians did too.
www.theguardian.com/australia-ne...

We’re losing species we love & major parties are sleepwalking through it.
Australians care. It’s time politicians did too.
www.theguardian.com/australia-ne...
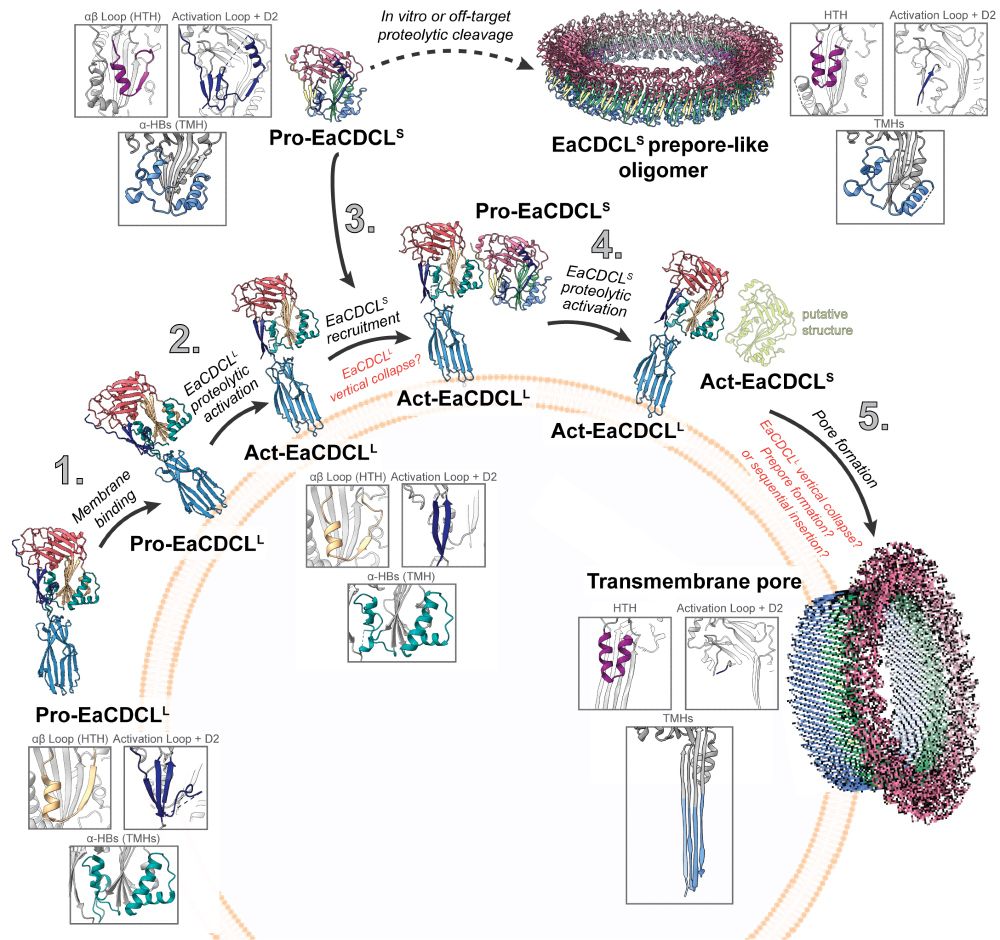
💡If you're passionate about mitochondrial research or know someone who is, please share! Apply now: careers.pageuppeople.com/513/cw/en/jo...
💡If you're passionate about mitochondrial research or know someone who is, please share! Apply now: careers.pageuppeople.com/513/cw/en/jo...
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Excited to see what people build now that "inject a llama with some junk and bleed it a month later" is no longer the cutting-edge tech
www.nature.com/articles/s41...
Excited to see what people build now that "inject a llama with some junk and bleed it a month later" is no longer the cutting-edge tech
www.nature.com/articles/s41...
This is such a cool initiative! DL4Proteins is democratizing deep learning for protein design and prediction at a pivotal time in science—a fantastic step toward making advanced protein engineering accessible to all!
github.com/Graylab/DL4P...
This is such a cool initiative! DL4Proteins is democratizing deep learning for protein design and prediction at a pivotal time in science—a fantastic step toward making advanced protein engineering accessible to all!
github.com/Graylab/DL4P...
www.biorxiv.org/content/10.1...
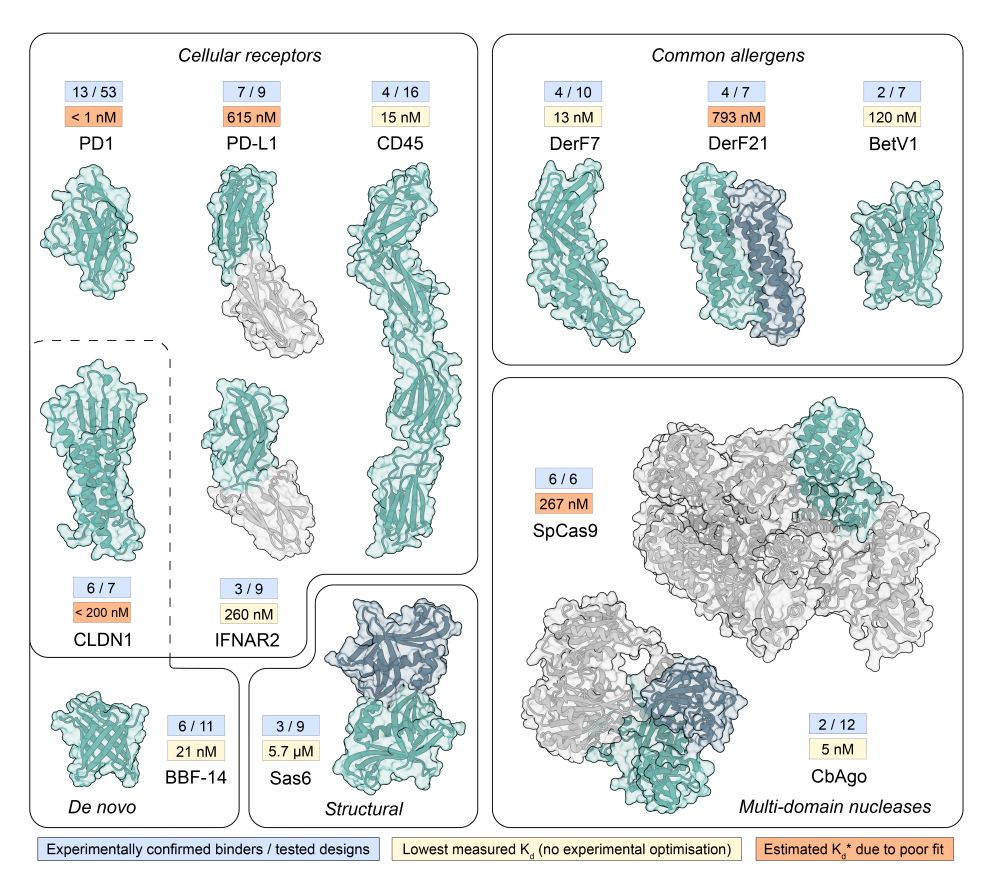
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
#CRISPR

www.biorxiv.org/content/10.1...
#CRISPR

